Abstract
Background:
The global pandemic of coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). About 18.4% of total Covid-19 cases were reported in children. Even though vertical transmission from mother to infant is likely to occur at a low rate, exposure to COVID-19 during fetal life may alter DNA methylation patterns with potential long-term effects.
Objective:
To determine if COVID-19 infection during pregnancy alters the DNA methylation patterns in umbilical cord blood cells from term infants and to identify potential pathways and genes affected by exposure to COVID-19 infection.
Methods:
Umbilical cord blood was collected from 8 infants exposed to COVID-19 during pregnancy and 8 control infants with no COVID-19 exposure. Genomic DNA was isolated from umbilical cord blood cells and genome-wide DNA methylation was performed using Illumina Methylation EPIC Array.
Results:
119 differentially methylated loci were identified at the FDR level of 0.20 (64 hypermethylated loci and 55 hypomethylated loci) in umbilical cord blood cells of COVID-19 exposed neonates compared to the control group. Important canonical pathways identified by Ingenuity Pathway Analysis (IPA) were related to stress response (corticotropin releasing hormone signaling, glucocorticoid receptor signaling, and oxytocin in brain signaling pathway), and cardiovascular disease and development (nitric oxide signaling in the cardiovascular system, apelin cardiomyocyte signaling pathways, factors promoting cardiogenesis, and renin-angiotensin signaling). The genes affected by the differential methylations were associated with cardiac, renal, hepatic, neurological diseases, developmental and immunological disorders.
Conclusions:
COVID-19 induces differential DNA methylation in umbilical cord blood cells. The differentially methylated genes may contribute to hepatic, renal, cardiac, developmental and immunological disorders in offspring born to mothers with COVID-19 infection during pregnancy, and their developmental regulation.
Keywords: Global DNA methylation, perinatal COVID-19, SARS-CoV-2, children, epigenetics, cord blood
Introduction
The United States (U.S.) has had over 1 million total deaths attributed to coronavirus disease (SARS-CoV-2) 2019 (COVID-19) since the beginning of the pandemic. 1 This is partly due to the highly infectious nature of SARS-CoV-2 compared to other coronavirus strains. The spike protein subunits S1 and S2, allow for the highly effective binding between the virus and host cells, after which the virus uses the host’s splicing machinery for its replication. 2 Even after widespread infection and now vaccination, it is likely that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) will remain present and become endemic in our world. 3 The pediatric population has also experienced a significant death toll as complications like multisystem inflammatory syndrome in children (MIS-C) have been well-documented. 4 Neonates are a vulnerable subgroup with a pressing need for a more detailed understanding of the effects of COVID-19.
While vertical transmission from mother to infant is likely to occur at a low rate, 5 active disease in neonates can still be severe. 6 The recent multinational study, INTERCOVID, described an increased rate of fetal distress, preterm birth, and lower birth weight in mothers with COVID-19 compared to controls. 7 Adverse perinatal outcomes from maternal COVID exposure likely relates to placental pathological thrombotic, vascular, and inflammatory changes documented in the setting of maternal COVID infection.8-11 Maternal systemic inflammatory response as well as inflammatory changes in the placenta can provoke a fetal inflammatory response, immune dysregulation, and epigenetic changes that could potentially have long-term consequences in offspring. 7 More recent studies have shown that COVID-19 infection during pregnancy can have an impact on the neonatal immune system affecting T cells and inducing unique inflammatory responses in the maternal-fetal environment.12,13 We also reported differential gene expression in umbilical cord blood (UCB) of infants exposed to COVID-19 infection during pregnancy. 14 These effects could have long-term impacts on the lives of offspring exposed to this environment, as has been historically seen with life-course studies from those exposed in-utero during the 1918 influenza pandemic. 15
The link between the perinatal environment and epigenetic changes that may affect disease has been documented in several studies.16,17 DNA methylation is one mechanism by which this change may occur. 18 It is an enzyme-mediated change during which a methyl group is added to cytosine at specific sites that leads to transcriptional regulation. 19 Intrauterine infections, such as histologic chorioamnionitis, have been associated with epigenetic changes via DNA methylation of neonatal genes.18,20 Exposure to COVID-19 during pregnancy is likely to have a similar effect in the perinatal environment that could lead to alteration in fetal DNA methylation. We hypothesized that early epigenetic signatures of COVID-19 exposure during pregnancy are detectable in UCB cells. The detection of DNA methylation changes at birth will help to identify at-risk offspring that can benefit from close monitoring and follow-up. Our objective for this study was to evaluate the DNA methylation patterns of human UCB cells from neonates exposed to COVID-19 in-utero and compare those with controls to better understand the epigenetic mechanisms associated with exposure to this virus during pregnancy and their developmental regulation.
Methods
Thomas Jefferson University Institutional Review Board (IRB) approved the study. The details of study design, informed consents, UCB sample collection, processing, storage, DNA isolation and global DNA methylation analysis is described elsewhere.14,18 In brief, UCB samples were collected from 8 infants exposed to COVID-19 infection during pregnancy and 8 control infants. The genome-wide DNA methylation study was performed using the Illumina Methylation EPIC Array (cat# WG-317-1001, Illumina Inc., San Diego, CA). DNA methylation loci with maximum beta values less than 0.001 across all the 16 samples (223 loci out of 865 918 loci) were filtered out from downstream analyses. Quantile normalization was used to normalize the beta values of remaining loci (865 695 loci). 21 Empirical Bayes method from the Limma package in statistical analysis software R version 4.1.2 was used for comparing the DNA methylation level differences between the Control group and the Covid-19 group. 22 The Benjamini and Hochberg method 23 was used to adjust the raw p-values to control for the false discovery rate (FDR) at 20%. We identified 119 DNA methylation loci at the FDR level of 0.20. Volcano plot was used to examine the association of fold change on log2 scale and the adjusted p-values. Heatmap was used to show the DNA methylation levels of significant DNA methylation loci. The clustering of DNA methylation loci and the samples were based on the ward.D2 clustering method according to Euclidean distance.
Results
Sixteen infants (8 infants COVID-19 group, 8 infants Control group) included in the study and genome-wide DNA methylation was performed on UCB DNA. Eight infants born either before the pandemic (n = 6) or maternal COVID-19 antibody negative at the time of delivery (n = 2) were included in the Control group. One mother was infected during the first trimester, 3 mothers during the second trimester and 4 mothers during the third trimester. Two mothers were symptomatic at the time of delivery, one with severe infection. None of the mothers in Control and COVID-19 groups received COVID-19 vaccine before the sample collection. All 16 infants were healthy at birth, there were no significant differences in mean birth weight (3.14 ± 0.63 kg vs 2.98 ± 0.56, P = .6), mean gestational age (38.1 ± 1.3vs 38.5 ± 1.4, P = .6) and other demographic and clinic characteristics in 2 groups described in detail elsewhere. 14
Differential DNA methylation
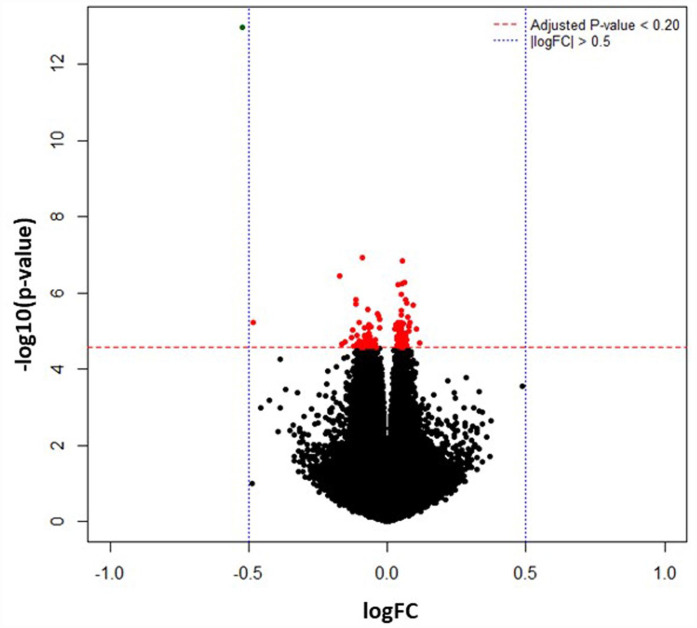
DNA methylation levels are measured by β-values which is the ratio of the methylated probe signal intensity to the total locus intensity. The β-values range from 0 (unmethylated) to 1 (fully methylated). DNA methylation loci with maximum beta values less than 0.001 across all the 16 samples (223 loci out of 865 918 loci) were filtered out from downstream analyses. The beta values of remaining loci (865 695 loci) were normalized using quantile normalization method. A total of 119 differentially methylated loci were identified, 64 hypermethylated loci (40 annotated genes) and 55 hypomethylated loci (38 annotated genes) (Supplemental Table 1). The top 20 hypo and hypermethylated gene names and probe-IDs based on p-values are listed in Tables 1 and 2. A volcano plot (Figure 1) was used to examine the association of fold change on log2 scale and the adjusted P-values.
Table 1.
Top 20 hypermethylated CpGs associated with Covid-19 infection during pregnancy.
| Target ID | Address A ID | Genome build | Reference Gene Name | Log Fold Change | t-statistics | P-value |
|---|---|---|---|---|---|---|
| cg24801716 | 81747151 | 37 | XYLB | 0.0519 | 9.048 | 1.46E-07 |
| cg14362073 | 16738900 | 37 | TBC1D9B | 0.0623 | 8.195 | 5.25E-07 |
| cg01270221 | 7662139 | 37 | SCD | 0.0398 | 8.119 | 5.91E-07 |
| cg06080729 | 98797987 | 37 | CLDN6 | 0.0483 | 7.725 | 1.10E-06 |
| cg10547050 | 70623213 | 37 | PHF12 | 0.0498 | 6.987 | 3.75E-06 |
| cg10308265 | 4802386 | 37 | UBTF | 0.0729 | 6.918 | 4.22E-06 |
| cg14002578 | 20636569 | 37 | GALNT12 | 0.0793 | 6.712 | 6.03E-06 |
| cg10463831 | 92740155 | 37 | KIF17 | 0.0390 | 6.701 | 6.15E-06 |
| cg05409441 | 4617426 | 37 | WDR90 | 0.0491 | 6.701 | 6.15E-06 |
| cg05473018 | 60616834 | 37 | SAMD14 | 0.0561 | 6.674 | 6.45E-06 |
| cg13422555 | 49738919 | 37 | CLK2 | 0.0321 | 6.649 | 6.73E-06 |
| cg03347381 | 26665135 | 37 | KIAA0226 | 0.0515 | 6.585 | 7.53E-06 |
| cg01998785 | 5793285 | 37 | LPCAT2 | 0.0788 | 6.577 | 7.64E-06 |
| cg00688330 | 41771897 | 37 | HJURP | 0.0509 | 6.488 | 8.94E-06 |
| cg13208845 | 88603875 | 37 | KNDC1 | 0.0271 | 6.478 | 9.10E-06 |
| cg11410237 | 87781944 | 37 | STRN4 | 0.0790 | 6.405 | 1.04E-05 |
| cg25052933 | 65752852 | 37 | HLA-DPB1 | 0.0610 | 6.345 | 1.15E-05 |
| cg07262630 | 70750521 | 37 | PLEC | 0.0525 | 6.274 | 1.31E-05 |
| cg15985126 | 23687174 | 37 | SAR1B | 0.0661 | 6.259 | 1.35E-05 |
| cg08648882 | 82735379 | 37 | ENSA | 0.0526 | 6.253 | 1.36E-05 |
Table 2.
Top 20 hypomethylated CpGs associated with Covid-19 infection during pregnancy.
| Target ID | Address A ID | Genome build | UCSC Reference Gene Name | Log Fold Change | t-statistics | P-value |
|---|---|---|---|---|---|---|
| cg20336007 | 65615162 | 37 | BMP7 | −0.5219 | −24.259 | 1.05E-13 |
| cg17377263 | 44735271 | 37 | RHOBTB1 | −0.0895 | −9.210 | 1.16E-07 |
| cg10000867 | 14737981 | 37 | CFAP69 | −0.1134 | −7.550 | 1.47E-06 |
| cg26700469 | 3675495 | 37 | WWOX | −0.1146 | −7.360 | 2.00E-06 |
| cg27134827 | 43688855 | 37 | PPT2 | −0.0345 | −7.039 | 3.43E-06 |
| cg14515812 | 40701105 | 37 | RAP1GAP | −0.0320 | −6.952 | 3.98E-06 |
| cg25587223 | 45650134 | 37 | GUCY2D | −0.0279 | −6.816 | 5.03E-06 |
| cg22931151 | 6643305 | 37 | OR2V2 | −0.4824 | −6.718 | 5.97E-06 |
| cg11275630 | 19766117 | 37 | SLC8A2 | −0.1028 | −6.708 | 6.08E-06 |
| cg17431391 | 56774849 | 37 | ZNF540 | −0.0674 | −6.639 | 6.85E-06 |
| cg03477130 | 20740100 | 37 | HLA-DOB | −0.0587 | −6.566 | 7.80E-06 |
| cg14119337 | 85685313 | 37 | MEG3 | −0.0689 | −6.540 | 8.15E-06 |
| cg02931058 | 76603468 | 37 | KLHL30 | −0.1105 | −6.263 | 1.34E-05 |
| cg20391833 | 42771267 | 37 | RPS6KA2 | −0.0791 | −6.219 | 1.45E-05 |
| cg25150313 | 95731891 | 37 | CDH23 | −0.0621 | −6.158 | 1.62E-05 |
| cg20952167 | 70682845 | 37 | MEG3; MEG3; MEG3 | −0.0647 | −6.141 | 1.67E-05 |
| cg11315777 | 79690504 | 37 | MAP3K7CL | −0.0726 | −6.140 | 1.67E-05 |
| cg26314066 | 87614203 | 37 | CNTD1 | −0.0418 | −6.132 | 1.70E-05 |
| cg12590430 | 2622447 | 37 | AMPH | −0.0578 | −6.122 | 1.73E-05 |
| cg10440011 | 78779322 | 37 | DPH1 | −0.0840 | −6.100 | 1.80E-05 |
Figure 1.
The differentially methylated CpG sites between the COVID-19 and the Control groups are depicted in this volcano plot. The x-axis shows difference in mean percent of DNA methylation and the y-axis shows the significance of differential methylation of each probe, represented as the negative log of the p-value. The red horizontal line indicates genome-wide significance threshold of FDR ⩽ 0.20. The blue vertical line signifies │Log Fold Change│>0.05.
Cluster analysis—Heatmap
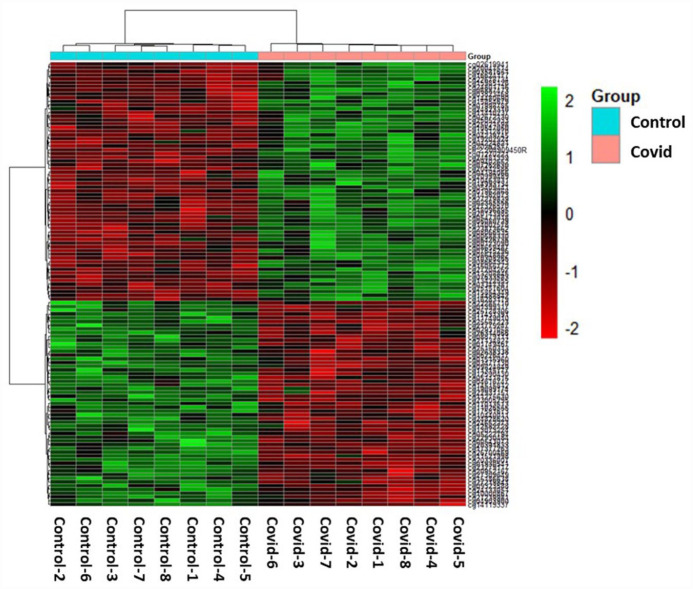
Hierarchical clustering method was used to perform the cluster analysis on the 119 differentially methylated loci for the 2 groups. Heatmap was used to show the DNA methylation levels of significant DNA methylation loci (Figure 2). The clustering of DNA methylation loci and the samples were based on the ward.D2 clustering method according to Euclidean distance.
Figure 2.
The 119 differentially methylated probe sets (FDR P-value ⩽ .20)) in the individual Covid-19 samples (right) compared to Control samples (left). The green and red colors indicate hyper-methylated and hypo-methylated loci, respectively.
Ingenuity pathway analysis
Ingenuity Pathway Analysis (IPA) software (https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis, Qiagen Inc., Germantown, MD) 24 was used for pathway analysis by loading 412 probe sets (290 annotated genes) that were differentially methylated with exposure to Covid-19 infection (P-value ⩽ .0001). A total of 340 canonical pathways were altered with exposure to Covid-19 infection during pregnancy. Selected key pathways important in Covid-19 pathophysiological response are shown in Table 3. Seventy-six diseases and functions were modified with Covid-19 exposure. Selected key diseases and functions altered with Covid-19 exposure are listed in Table 4.
Table 3.
Canonical pathways picked-up by ingenuity pathway analysis of the differentially methylated genes in Covid-19 group compared to the Control group.
| Ingenuity Canonical Pathways | -log (P-value) | Genes involved in pathways | Importance |
|---|---|---|---|
| Nitric Oxide Signaling in the Cardiovascular System | 3.95 | CACNG8,GUCY2D,ITPR2, NOS3,PIK3CB,PRKCE | Cardiovascular and immune systems |
| Corticotropin Releasing Hormone Signaling | 3.35 | CACNG8,CREB5,GUCY2D, ITPR2,NOS3,PRKCE | Pituitary gland, stress response |
| Factors Promoting Cardiogenesis in Vertebrates | 3.34 | BMP3,BMP7,CER1,CREB5, PRKCE,WNT6 | Formation of heart in neonates |
| Apelin Cardiomyocyte Signaling Pathway | 3.31 | MAPK6,NOS3,PIK3CB, PRKCE,SLC8A2 | Cardiovascular diseases, Obesity and Cancer |
| Oxytocin Signaling Pathway | 3.3 | CACNG8,CREB5,GUCY2D, ITPR2,MAPK6,NOS3, PIK3CB,PRKCE | Promotes lactation, involved in reproductive and social behavior |
| Growth Hormone Signaling | 2.9 | IGF1R,PIK3CB,PRKCE, RPS6KA2 | Stimulate the pituitary gland, control metabolism and growth |
| Estrogen Receptor Signaling | 2.85 | CACNG8,CREB5,FOXA1, IGF1R,NOS3,PIK3CB, PPARGC1A,PRKCE,TRRAP | Promotes sexual organ maturation and development in female |
| Endothelin-1 Signaling | 2.82 | GUCY2D,ITPR2,MAPK6, NOS3,PIK3CB,PRKCE | Vasoconstrictive action, causes fibrosis of the vascular cells |
| Synaptic Long Term Depression | 2.8 | CACNG8,GUCY2D,IGF1R, ITPR2,NOS3,PRKCE | Weakens synapses efficiency |
| Adrenomedullin signaling pathway | 2.73 | GPR182,GUCY2D,ITPR2, MAPK6,NOS3,PIK3CB | Regulation of endothelial barrier function, circulation control, vasodilation |
| Insulin Secretion Signaling Pathway | 2.69 | CACNG8,CREB5,ITPR2,PCSK2,PIK3CB,PRKCE,SEC11A | Crucial role on cell metabolism, growth, proliferation and differentiation |
| Opioid Signaling Pathway | 2.6 | CACNG8,CREB5,ITPR2, MAPK6,NOS3,PRKCE, RPS6KA2 | Mediates analgesic effect |
| Role of NFAT in Cardiac Hypertrophy | 2.47 | CACNG8,IGF1R,ITPR2, PIK3CB,PRKCE,SLC8A2 | Development of the cardiovascular system, Embryonic heart |
| eNOS Signaling | 2.47 | HSPA4,ITPR2,NOS3, PIK3CB,PRKCE | Vascular homeostasis, contributes to angiogenesis |
| AMPK Signaling | 2.25 | CREB5,NOS3,PIK3CB,PPARGC1A,PPM1H,SMARCE1 | Regulation of cellular energy homeostasis |
| Renin-Angiotensin Signaling | 2.07 | ITPR2,PAK2,PIK3CB,PRKCE | Controls blood pressure and sodium homeostasis, critical regulator of blood volume and systemic vascular resistance |
| Oxytocin In Brain Signaling Pathway | 2.03 | CACNG8,CREB5,ITPR2, PIK3CB,PRKCE | Significant roles in heart and brain, regulates hypothalamo-pituitary-adrenal axis, modulating behavioral response toward stress and social behavior |
| FXR/RXR Activation | 2.02 | FOXA1,PPARGC1A,RBP4, SLCO1B3 | Alters expression of different groups of genes involved in bile acid homeostasis, lipid metabolism, and glucose balance |
| Angiopoietin Signaling | 1.85 | NOS3,PAK2,PIK3CB | Involved in vascular development and stability, angiogenesis and quiescence |
| Glucocorticoid Receptor Signaling | 1.56 | DNAH10,HLA-DOB,HLA-DPB1, HSPA4,IL1R2,NOS3, PIK3CB,SMARCE1 | plays essential role in the cardiovascular system |
| Circadian Rhythm Signaling | 1.51 | CACNG8,CREB5,GUCY2D, ITPR2,PRKCE | regulates the circadian rhythm of a cell or organism, senses external timing signals like light/dark |
| Th1 Pathway | 1.37 | HLA-DOB, HLA-DPB1, PIK3CB | Mediates cellular response to infection, identifies and eradicates of intracellular pathogens such as viruses and bacteria |
Table 4.
Modified functions and diseases obtained by differentially methylated genes after exposure to Covid-19 infection during pregnancy.
| Modified diseases and functions | Range of P-values for genes involved | Number of genes involved |
|---|---|---|
| Cancer | 3.98E-07-3.9E-02 | 138 |
| Organismal Injury and Abnormalities | 3.98E-07-3.9E-02 | 140 |
| Dermatological Diseases and Conditions | 9.24E-06-1.97E-02 | 108 |
| Gastrointestinal Disease | 2.82E-05-3.72E-02 | 128 |
| Cell Morphology | 1.3E-04-3.9E-02 | 17 |
| Hepatic System Disease | 1.8E-04-1.97E-02 | 83 |
| Reproductive System Disease | 3.27E-04-3.9E-02 | 95 |
| Cellular Development | 4.28E-04-3.83E-02 | 24 |
| Connective Tissue Development and Function | 4.28E-04-3.9E-02 | 10 |
| Tissue Morphology | 4.28E-04-3.9E-02 | 15 |
| Endocrine System Disorders | 5.23E-04-3.9E-02 | 118 |
| Embryonic Development | 6.4E-04-3.26E-02 | 12 |
| Organismal Development | 6.4E-04-3.9E-02 | 17 |
| Cellular Growth and Proliferation | 7.14E-04-3.9E-02 | 24 |
| Nervous System Development and Function | 7.14E-04-3.83E-02 | 9 |
| Neurological Disease | 8.91E-04-3.9E-02 | 92 |
| Developmental Disorder | 1.03E-03-3.9E-02 | 29 |
| Renal and Urological Disease | 1.03E-03-3.63E-02 | 14 |
| Cardiovascular Disease | 1.4E-03-2.36E-02 | 5 |
| Cell-To-Cell Signaling and Interaction | 1.91E-03-3.9E-02 | 16 |
| Hematological System Development and Function | 1.91E-03-3.9E-02 | 14 |
| Immunological Disease | 3.23E-03-3.61E-02 | 43 |
| Respiratory Disease | 5.78E-03-3.9E-02 | 63 |
Tox functions
COVID-19 exposure during pregnancy potentially altered 26 tox functions in UCB (Table 5). Top tox functions related to cardiotoxicity include cardiac arteriopathy, cardiac stenosis, congenital heart anomaly. Top altered hepatotoxic functions are liver hyperbilirubinemia, liver proliferation, liver inflammation/hepatitis, liver cirrhosis, liver fibrosis, liver hyperplasia/hyperproliferation, and liver cholestasis. Top altered tox functions related to nephrotoxicity are nephrosis, renal dysplasia, renal hypoplasia, renal necrosis/cell death, renal proliferation, glomerular injury, renal inflammation, renal nephritis.
Table 5.
Top tox functions modified with Covid-19 infection during pregnancy.
| Modified Tox Functions | Category | Range of P-values for genes involved | Number of Molecules |
|---|---|---|---|
| Cardiac Arteriopathy | Cardiotoxicity | 6.61E-03-2.97E-01 | 4 |
| Cardiac Stenosis | 8.26E-02-8.26E-02 | 1 | |
| Congenital Heart Anomaly | 1.01E-01-4.07E-01 | 2 | |
| Liver Hyperplasia/Hyperproliferation | Hepatotoxicity | 2.65E-04-5.5E-01 | 74 |
| Liver Hyperbilirubinemia | 1.97E-02-1.97E-02 | 1 | |
| Liver Proliferation | 8.87E-02-8.87E-02 | 1 | |
| Liver Inflammation/Hepatitis | 2.2E-01-4.35E-01 | 2 | |
| Liver Cirrhosis | 2.43E-01-5.28E-01 | 3 | |
| Liver Fibrosis | 2.43E-01-5.28E-01 | 3 | |
| Liver Cholestasis | 3.33E-01-3.33E-01 | 1 | |
| Nephrosis | Nephrotoxicity | 6.61E-03-6.61E-03 | 1 |
| Renal Dysplasia | 6.61E-03-7.62E-03 | 2 | |
| Renal Hypoplasia | 6.61E-03-6.61E-03 | 1 | |
| Renal Necrosis/Cell Death | 3.3E-02-5.41E-01 | 7 | |
| Renal Proliferation | 1.44E-01-1.44E-01 | 3 | |
| Glomerular Injury | 2.23E-01-5.44E-01 | 2 | |
| Renal Inflammation | 2.23E-01-5.44E-01 | 2 | |
| Renal Nephritis | 2.23E-01-5.44E-01 | 2 |
Upstream regulators
Upstream regulator analysis by IPA identified 72 regulators to be dysregulated. Key identified regulators and their target molecules are described in Table 6.
Table 6.
Key upstream regulators dysregulated with Covid-19 exposure during pregnancy.
| Upstream Regulators | Molecule Type | P-value of overlap | Target Molecules in Dataset |
|---|---|---|---|
| LPIN1 | phosphatase | .000441 | LPIN1,PPARGC1A |
| KLF5 | transcription regulator | .000508 | CAVIN1,FOXA1,NMI,PTGES |
| AREG | growth factor | .000851 | CCNF,HJURP,KIF20A,SSRP1 |
| MITF | transcription regulator | .00112 | CCNF,KIF20A,KIFC1,PPARGC1A,SCD |
| HCAR2 | G-protein coupled receptor | .00282 | KIF20A,NMI |
| mir-218 | microRNA | .00443 | MDGA1,SASH1 |
| EPAS1 | transcription regulator | .0055 | HSPA4,LOX,NEK8,NOS3 |
| NPC2 | transporter | .00637 | CREB5,PTGES |
| Histone h3 | group | .00784 | FOXA1,GBX2,IGF1R,NOS3,PTGES,SCD |
| EZH2 | transcription regulator | .0125 | BMP7,CER1,FOXA1,mir-10,PTGES |
Discussion
In this observational and hypothesis generating study, we report that COVID-19 infection during pregnancy is associated with differential DNA methylation in UCB cells. Exposure to COVID-19 infection during pregnancy altered the fetal patterns of DNA methylation in genes related to stress response (corticotropin releasing hormone signaling, glucocorticoid receptor signaling, and oxytocin in brain signaling pathway), and cardiovascular disease and development (nitric oxide signaling in the cardiovascular system, factors promoting cardiogenesis, and apelin cardiomyocyte signaling pathways, and renin-angiotensin signaling). The genes affected by the differential methylations were associated with cardiac, renal, hepatic, neurological diseases, developmental and immunological disorders. The genes involved with dysregulated methylation were also associated with cardiac, hepatic, and renal toxicity and key up-stream regulators.
The top hypomethylated genes included BMP7 (Bone morphogenetic protein 7), RHOBTB1 (Rho related BTB domain containing 1), WWOX (WW domain containing oxidoreductase), and HLA-DOB (Major histocompatibility complex, class II, DO beta). BMP7 codes a ligand of TGF-beta (transforming growth factor-beta) that binds certain TGF-beta receptors and leads to recruitment and activation of SMAD family transcription factors. BMP7 is associated with bone disorders, renal injury, and pulmonary fibrosis.25-27 Progressive pulmonary fibrosis (lung remodeling) is thought to lead to higher mortality in COVID-19 patients. Severity of disease is associated with activation of the TGF-beta pathways that triggered inflammation, apoptosis, and fibrosis. The BMP7 pathway has been shown to oppose the negative effects of TGF-beta, blocking inflammation and apoptosis and reversing fibrosis. 28 Hypomethylation of BMP7 gene if persisted later in life can potentially reduce the risk of lung fibrosis in offspring. WWOX encodes a member of short-chain dehydrogenases/reductases protein family and appears to function as tumor suppressor gene. In a recent study, Dugan et al demonstrated that the genetic locus containing the WWOX gene was implicated as a clinical Alzheimer’s disease risk locus. 29 In 2019, a study reported the vital implications of WWOX in atherosclerosis and cardiovascular diseases. 30 Another study has shown that inhibition of WWOX, contributes to pulmonary remodeling and subsequent pulmonary arterial hypertension. 31 In a gene ontology study on Covid-19 patients with respiratory failure, Oh and coworkers identified WWOX to be one of the 8 genes from the largest connected network. 32 HLA-DOB suppresses peptide loading of MHC class II molecules. Recently, HLA-DOB was observed to be one of the 13 key differentially expressed genes in preterm infants who developed bronchopulmonary dysplasia (BPD) and expression was increased in infants with BPD. 33 Due to hypomethylation of HLA-DOB CpGs with COVID-19 infection in pregnancy, expression of HLA-DOB is likely to be increased in preterm infants and could potentially increase the risk of BPD.
The top hypermethylated genes included XYLB (Xylulose kinase), CLDN6 (Claudin-6), UBTF (Upstream binding transcription factor), LPCAT2 (Lysophosphatidylcholine acyltransferase 2), STRN4 (Striatin 4), and HLA-DPB1 (Major histocompatibility complex, class II, DP beta 1). XYLB plays an important role in the regulation of glucose metabolism and lipogenesis. Decreased plasma product from XYLB was hypothesized as one of the potential metabolomics biomarkers of COVID-19.34-36 CLDN6 (Claudin-6) has a large role in tight junction-specific obliteration of the intercellular space (which has a role in microbial infection), and also functions as a receptor for hepatitis C virus entry into hepatic cells. UBTF gene is associated with neurodegeneration, brain atrophy, intellectual disability, retinoblastoma, pediatric acute myeloblastic leukemia.37,38 Integrative network analysis in transcriptional landscape of circulating platelets from COVID-19 patients identified downregulated UBTF as a key factor for disease severity. Abate et al. has shown that LPCAT2 co-localizes with TLR4 and modulates macrophage inflammatory gene expression in response to lipopolysaccharide. 39 In another study LPCAT2 methylation was identified to be a biomarker for cedar pollen allergic rhinitis severity in the Japanese population. 40 STRN4 (Striatin 4) is associated with muscular dystrophy, cerebral cavernous malformations and cancer. Chakraborty et al and coworker have reported upregulation of STRN4 gene expression with Sars-Cov-2 infection. 41
We compared the differentially methylated loci with our recently published gene expression data. 14 Four genes are common for both DNA methylation and gene expression data. Two differentially methylated genes had reciprocal changes in gene expression. XYLB is hypermethylated with reciprocal downregulated gene expression and ras-associated protein-1 GTPase-activating-protein (RAP1GAP) is hypomethylated with reciprocal upregulated gene expression. In animal models, RAP1GAP inhibit autophagy, increase oxidative stress and mediates angiotensin II-induced cardiomyocyte hypertrophy. 42 Two other genes, HLA-DPB1 and ENSA have methylation and gene expression in the same direction (hypermethylated loci and upregulated gene expression).
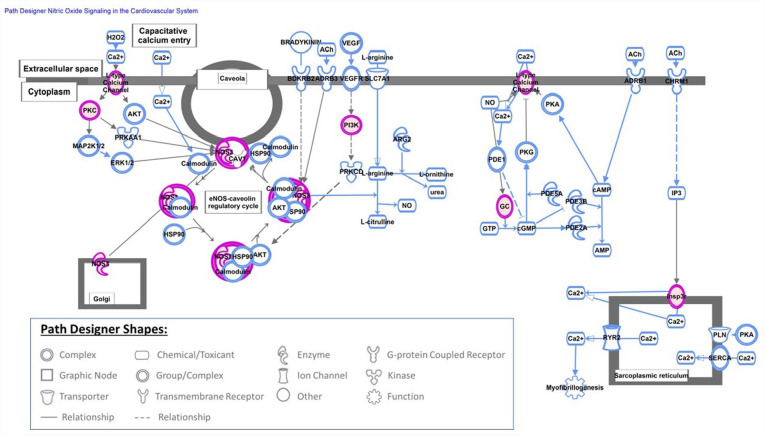
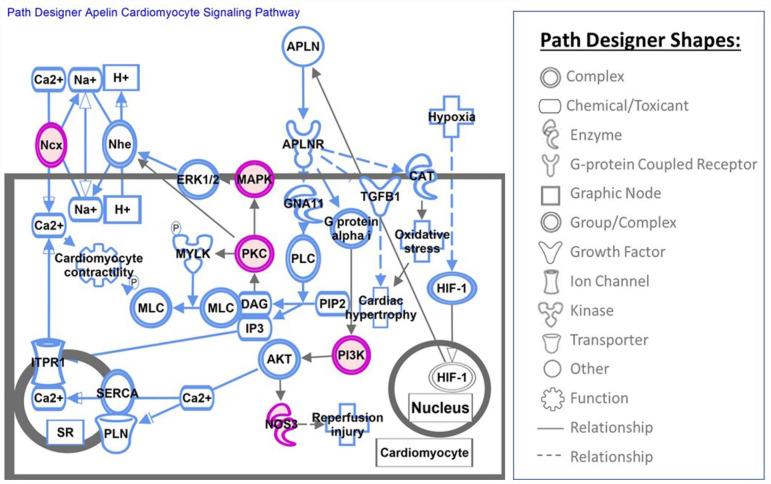
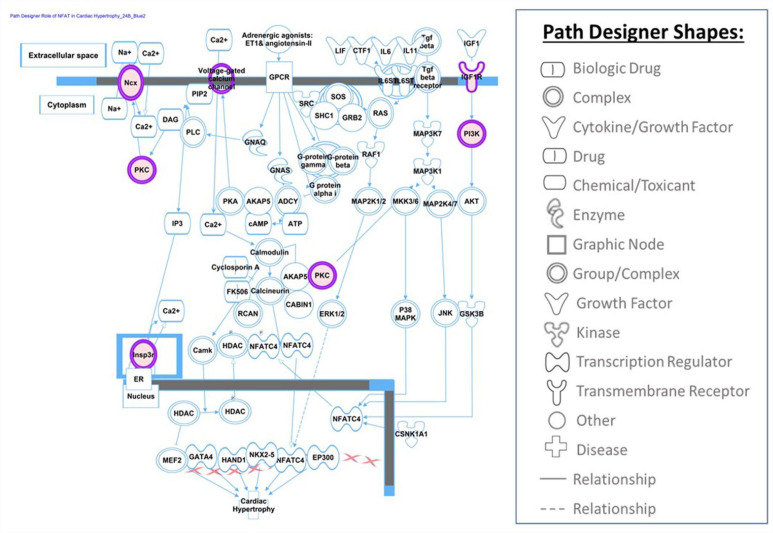
Altered methylation pattern in genes involved in canonical pathways can potentially link COVID-19 exposure during pregnancy to short- and long-term outcomes in offspring. At least, 340 canonical pathways were found to be altered in UCB cells exposed to COVID-19 infection during pregnancy. COVID-19 infection as well as stress exposure may alter patterns of DNA methylation in genes related to stress. Altered DNA methylation status of stress-related genes have been reported as the epigenetic changes due to adverse events during early life including fetal period.43-45 Our data also indicates that infection with COVID-19 in pregnancy altered the fetal patterns of DNA methylation in genes related to the stress response. The methylation of genes involved in corticotropin releasing hormone signaling, glucocorticoid receptor signaling, opioid signaling pathway and oxytocin in brain signaling pathway were affected by the exposure to COVID-19 infection. Persistence of altered DNA methylation in genes related to stress can potentially affect child development and increase the risk for behavior and psychological disorders. COVID-19 infection during pregnancy also altered the DNA methylation pattern in genes involved in cardiovascular development and disease. The most altered canonical pathway in our cohort after exposure to COVID-19 infection was nitric oxide (NO) signaling in the cardiovascular system (Figure 3). Nitric oxide is a free radical that plays an important physiological role in cardiovascular and immune systems. 46 We also observed an altered DNA methylation pattern in genes related to: cardiogenesis in vertebrates, the apelin cardiomyocyte signaling pathway (Figure 4), NFAT (nuclear factor of activated T cells) and cardiac hypertrophy (Figure 5), and the eNOS signaling pathway. The role of NO, apelin, and NFAT have been reported in the pathogenesis of COVID-19 infection. 47 Apelin plays an important role in alleviating Ang-II mediated acute lung and cardiovascular injuries and prothrombotic events in COVID-19 patients. 48 The NFAT (nuclear factor of activated T cells) pathway contributes to the pathogenesis of cardiac hypertrophy and plays a critical role in Ca(2+)/calcineurin-mediated cardiac hypertrophic signaling. Excessive CaN/NFAT activation and TRPC overproduction in the heart contribute to the development and progression of pathological myocardial hypertrophy. 49 The role of altered gene methylation related to these pathways needs further investigation if persistence of altered methylation in infants increases the risk of heart disease later in life.
Figure 3.
Nitric oxide (NO) signaling in the cardiovascular system is shown in this figure which is the top-most canonical pathway picked up by Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., https://digitalinsights.qiagen.com/IPA) by loading 412 probe sets (290 annotated genes) that were found to be differentially methylated with exposure to COVID-19, P-value ⩽ .0001.
Figure 4.
This figure depicts the Apelin cardiomyocyte signaling canonical pathway obtained from IPA pathway analysis. Apelin is an important regulator of cardiac contractility in cardiomyocytes and also protects the heart against cardiac hypertrophy and ischemia-reperfusion injury.
Figure 5.
Role of NFAT in Cardiac Hypertrophy pathway is presented in this figure.
Exposure to COVID-19 infection during pregnancy altered 26 toxic functions in our cohort. The top toxic functions that altered DNA methylation were seen in genes related to hepatoxicity, cardiotoxicity, and nephrotoxicity. Cardiac, hepatic and renal dysfunctions are commonly observed in children with MIS-C. 50 Li et al reported that the SARS-CoV-2 virus can shut down energy production in cells of the kidneys, heart, spleen and other organs in an animal model. 51 Additionally, late cardiovascular, liver and renal complications after COVID-19 infection have been reported. 52 Altered DNA methylation in genes related to cardiac, hepatic and renal toxic functions after COVID-19 infection during pregnancy may potentially increase the risk for heart, liver and kidney diseases in offspring and should have longitudinal follow-up.
Upstream regulator analysis by IPA identified 72 regulators to be dysregulated after COVID-19 exposure during pregnancy including Krüppel-like factor 5 (KLF5), melanocyte inducing transcription factor (MITF), endothelial PAS domain protein 1 (EPAS1) and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2). KLF5 is a zinc finger-type transcription factor and is involved both in promoting and suppressing cell proliferation. KLF5 was associated with severe COVID-19 infection and hospitalization. 53 The diseases associated with KLF5 include: cardiomyopathy, sigmoid disease, familial partial lipodystrophy, arteriosclerosis and pancreatic cancer. 54 MITF plays a role in the development, survival, and function of various types of cells. In melanocytes, this protein controls production of melanin, which contributes to hair, eye, and skin color. Jeong et al identified MITF to be one of the transcription factors that could drive the transcriptomic changes for the pathogenesis of COVID-19 and markers related to its severity. 55 The EPAS1 gene codes for a transcription factor involved in the induction of genes regulated by oxygen, which is induced as hypoxia occurs. EPAS1 regulates vascular endothelial growth factor (VEGF) expression, which is involved in the development of blood vessels and the tubular system of lung. EZH2 provides instructions for making the histone methyltransferase enzyme that can suppress the activity of certain genes via methylation. LIPN1 (Lipin 1) acts as a magnesium-dependent phosphatidate phosphatase enzyme and is involved in adipocyte differentiation. The protein encoded by AREG (ampiregulin) gene is a member of the epidermal growth factor family. Using bulk RNA-sequencing showed an increase of AREG in the peripheral blood monocytes (PBMCs) of COVID-19 patients, supporting a potential role of AREG in SARS-CoV-2-induced lung pathology. 56 Bui et al have reported that AREG is significantly upregulated in PBMCs, CD4T Cells, NK cells, neutrophils, and dendritic cells, suggesting that upregulation of AREG may be an attempt to ameliorate the severe injury induced by SARS-CoV-2 infection. 57 HCAR2 (Hydroxycarboxylic Acid Receptor 2) is a niacin receptor associated with inflammatory bowel disease, diversion colitis and pellagra. NPC2 (NPC Intracellular Cholesterol Transporter 2) is secreted from the liver into bile and plasma, where it may have a functional role in cholesterol transport in physiologic and pathologic conditions. 58 In a 2021 study, Prasad et al reported that NPC2 is one of the genes linked with neurological disease, which is also connected to COVID-19. 59 Histone H3 is one of the 5 main histones involved in the structure of chromatin in eukaryotic cells. Histone H3 plays a crucial role in activating the spindle assembly checkpoint in response to a defect in mitosis. The presence of histone H3 during ICU stays was associated with thromboembolic events and secondary infection. 58 Non-cleaved histone H3 was associated with the need for vasoactive treatment, invasive ventilation, and the development of acute kidney injury in COVID-19 patients. 60 In a review article, Ligi et al. concluded that monitoring systemic histone concentrations (both unmodified and citrullinated histone H3) of patients with SARS-CoV-2 infection throughout their hospitalization (especially in critical patients and/or those needing intensive care) may be a useful tool for early prediction of higher risk of disease progression. 61
To our knowledge, this is the first study reporting differential DNA methylation in UCB cells from infants exposed to COVID-19 infection during pregnancy. More recently, Hill et.al, reported differential methylation of several genes including genes in pathways that play a role in human neurodevelopment in buccal swabs on infants exposed to in utero COVID-19 infection. 62 The altered methylation pattern identified in genes related to hepatic, renal, and cardiac diseases show a potential connection to long-term consequences of COVID-19 infection. Our study, however, also has several limitations, for example, the sample size of 16 neonates is small, but similar limited sample sizes have been used in studies investigating differential DNA methylation patterns.63,64 There is an inherent chance of finding differences in the DNA methylation due to multiple comparison and our results should be considered as being hypothesis generating and should be validated in a larger cohort.
In conclusion, SARS-CoV-2 COVID-19 exposure during pregnancy induces differential DNA methylation in UCB cells. We identified 119 differentially methylated genes, important tox functions, upstream regulators and canonical pathways related to the stress response, hepatotoxicity, nephrotoxicity and cardiotoxicity in UCB cells of neonates born to mothers with COVID-19 infection during pregnancy. Future studies can further validate differential methylation of target genes in a larger cohort of infants. Our data may help to further understanding the role of key pathways and genes on the long-term consequences related to COVID-19 infection during pregnancy. Functional studies on the identified pathways and genes could lead to the development of biomarkers for the illnesses in offspring caused COVID-19 infection during pregnancy. Lastly, our results underscore the importance of determining downstream consequences in offspring of mothers who had COVID-19 infection during pregnancy even in the absence of vertical transmission of disease.
Supplemental Material
Supplemental material, sj-docx-1-gae-10.1177_25168657231184665 for SARS-CoV-2 Covid-19 Infection During Pregnancy and Differential DNA Methylation in Human Cord Blood Cells From Term Neonates by Pedro Urday, Suhita Gayen nee’ Betal, Rochelle Sequeira Gomes, Huda B Al-Kouatly, Kolawole Solarin, Joanna SY Chan, Dongmei Li, Irfan Rahman, Sankar Addya, Rupsa C Boelig and Zubair H Aghai in Epigenetics Insights
Acknowledgments
The authors acknowledge the support of Cancer Genomics Facility, Sidney Kimmel Cancer Center, Thomas Jefferson University, Philadelphia. Differential gene expression data from the same cohort is recently published by our group. 14
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study funded in part through Pilot Grant (ZA) through an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number U54-GM104941 (PI: Hicks), an NICHD grant 3R21HD101127-01S1 (PI RCB) and Hemant Desai Fellowship Award (PU).
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contribution Statement: PU, SGB contributed equally to concept and design, sample collection and processing, acquisition and assembly of data, data analysis and interpretation and manuscript writing and share first authorship. SA contributed to data analysis and interpretation. RSG, JSC, HA, RCB, KS, DL, IR contributed to concept and design, sample collection and processing, and manuscript writing. ZA contributed to concept and design, data analysis and interpretation and manuscript writing. All authors have approved the version of the submitted manuscript.
ORCID iD: Zubair H. Aghai  https://orcid.org/0000-0001-5545-9706
https://orcid.org/0000-0001-5545-9706
Supplemental material: Supplemental material for this article is available online.
References
- 1.Centers for Disease Control and Prevention. COVID data tracker. Accessed January 18, 2023. https://covid.cdc.gov/covid-data-tracker
- 2.Li T, Huang T, Guo C, et al. Genomic variation, origin tracing, and vaccine development of SARS-CoV-2: A systematic review. Innovation. 2021;2:100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marks P, Woodcock J, Califf R. COVID-19 vaccination-becoming part of the new normal. JAMA. 2022;327:1863-1864. [DOI] [PubMed] [Google Scholar]
- 4.Nakra NA, Blumberg DA, Herrera-Guerra A, Lakshminrusimha S. Multi-system inflammatory syndrome in children (MIS-C) following SARS-CoV-2 infection: review of clinical presentation, hypothetical pathogenesis, and proposed management. Children. 2020;7(7):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massalha M, Yefet E, Rozenberg O, et al. Vertical transmission and humoral immune response following maternal infection with SARS-CoV-2: a prospective multicenter cohort study. Clin Microbiol Infect. 2022;28:1258-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahfouz ME, Elrewiny M, Abdel-Moneim AS. Clinical manifestations of SARS-CoV-2 infection in neonates and the probability of maternal transmission. J Paediatr Child Health. 2022;58:1366-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giuliani F, Oros D, Gunier RB, et al. Effects of prenatal exposure to maternal COVID-19 and perinatal care on neonatal outcome: results from the INTERCOVID multinational cohort study. Am J Obstet Gynecol. 2022;227:488.e1-488.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prochaska E, Jang M, Burd I. COVID-19 in pregnancy: placental and neonatal involvement. Am J Reprod Immunol. 2020;84:e13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penfield CA, Brubaker SG, Limaye MA, et al. Detection of severe acute respiratory syndrome coronavirus 2 in placental and fetal membrane samples. Am J Obstet Gynecol MFM. 2020;2:100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharps MC, Hayes DJL, Lee S, et al. A structured review of placental morphology and histopathological lesions associated with SARS-CoV-2 infection. Placenta. 2020;101:13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boelig RC, Aghai ZH, Chaudhury S, Kazan AS, Chan JSY, Bergmann-Leitner E. Impact of COVID-19 disease and COVID-19 vaccination on maternal or fetal inflammatory response, placental pathology, and perinatal outcomes. Am J Obstet Gynecol. 2022;227:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Flores V, Romero R, Xu Y, et al. Maternal-fetal immune responses in pregnant women infected with SARS-CoV-2. Nat Commun. 2022;13:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gee S, Chandiramani M, Seow J, et al. The legacy of maternal SARS-CoV-2 infection on the immunology of the neonate. Nat Immunol. 2021;22:1490-1502. [DOI] [PubMed] [Google Scholar]
- 14.Gayen Nee’ Betal S, Urday P, Al-Kouatly HB, et al. COVID-19 infection during pregnancy induces differential gene expression in human cord blood cells from term neonates. Front Pediatr. 2022;10:834771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almond D, Mazumder B. The 1918 Influenza pandemic and subsequent Health Outcomes: an Analysis of SIPP data. Fam Econ Rev. 2005;95:258-262. [DOI] [PubMed] [Google Scholar]
- 16.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412-417. [DOI] [PubMed] [Google Scholar]
- 17.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297-307. [DOI] [PubMed] [Google Scholar]
- 18.Fong G, Gayen Nee’ Betal S, Murthy S, et al. DNA methylation profile in human cord blood mononuclear leukocytes from term neonates: Effects of histological chorioamnionitis. Front Pediatr. 2020;8:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacol. 2013;38:23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Hoyo C, Murphy S, et al. DNA methylation at imprint regulatory regions in preterm birth and infection. Am J Obstet Gynecol. 2013;208:395.e1-395.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics? 2003;19:185-193. [DOI] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289-300. [Google Scholar]
- 24.Krämer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics? 2014;30:523-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ural O, Kıratlı HE, Sümer, et al. [Evaluation of annexin-1 (ANXA-1), annexin-2 (ANXA-2) and bone morphogenetic protein-7 (BMP-7) serum levels in patients followed up with A diagnosis of COVID-19]. Mikrobiyol Bul. 2022;56:25-35. [DOI] [PubMed] [Google Scholar]
- 26.Raming R, Cordasic N, Kirchner P, et al. Neonatal nephron loss during active nephrogenesis results in altered expression of renal developmental genes and markers of kidney injury. Physiol Genomics. 2021;53:509-517. [DOI] [PubMed] [Google Scholar]
- 27.Liang D, Wang Y, Zhu Z, et al. BMP-7 attenuated silica-induced pulmonary fibrosis through modulation of the balance between TGF-β/Smad and BMP-7/Smad signaling pathway. Chem Biol Interact. 2016;243:72-81. [DOI] [PubMed] [Google Scholar]
- 28.Carlson FR, Jr., Bosukonda D, Keck PC, Carlson WD. Multiorgan damage in patients with COVID-19: is the TGF-β/BMP pathway the missing link? JACC Basic Transl Sci. 2020;5:1145-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dugan AJ, Nelson PT, Katsumata Y, et al. Association between WWOX/MAF variants and dementia-related neuropathologic endophenotypes. Neurobiol Aging. 2022;111:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teixeira Gomes M, Chen J, Haider S, Bai Y, Singla S, Machado RF. Smooth Muscle Cell Loss of the Tumor Suppressor WWOX Contributes to the Development of Pulmonary Hypertension. D106 PICK YOUR POISON: GENES, DRUGS, AND PAH. American Thoracic Society International Conference Abstracts: American Thoracic Society; 2019: A7211-A. [Google Scholar]
- 31.Tanna M, Aqeilan RI. Modeling WWOX loss of function in vivo: what have we learned? Front Oncol. 2018;8:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh JH, Tannenbaum A, Deasy JO. Identification of biological correlates associated with respiratory failure in COVID-19. BMC Med Genomics. 2020;13:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Wu M, Wang F, et al. CD74, a novel predictor for bronchopulmonary dysplasia in preterm infants. Med. 2020;99:e23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nahalka J. Theoretical analysis of S, M and N structural proteins by the Protein–RNA recognition code leads to genes/proteins that are relevant to the SARS-CoV-2 life cycle and Pathogenesis. Front Genet. 2021;12:763995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Silva IW, Nayek S, Singh V, Reddy J, Granger JK, Verbeck GF. Correction: Paper spray mass spectrometry utilizing Teslin® substrate for rapid detection of lipid metabolite changes during COVID-19 infection. Analyst. 2022;147:1515. [DOI] [PubMed] [Google Scholar]
- 36.Wu D, Shu T, Yang X, et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl Sci Rev. 2020;7:1157-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toro C, Hori RT, Malicdan MCV, et al. A recurrent de novo missense mutation in UBTF causes developmental neuroregression. Hum Mol Genet. 2018;27:691-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edvardson S, Nicolae CM, Agrawal PB, et al. Heterozygous De novo UBTF Gain-of-Function variant is associated with neurodegeneration in childhood. Am J Human Genet. 2017;101:267-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abate W, Alrammah H, Kiernan M, Tonks AJ, Jackson SK. Lysophosphatidylcholine acyltransferase 2 (LPCAT2) co-localises with TLR4 and regulates macrophage inflammatory gene expression in response to LPS. Sci Rep. 2020;10:10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe H, Miyake K, Matsuoka T, et al. LPCAT2 methylation, a novel biomarker for the severity of cedar pollen allergic rhinitis in Japan. Am J Rhinol Allergy. 2021;35:631-639. [DOI] [PubMed] [Google Scholar]
- 41.Chakraborty C, Sharma AR, Bhattacharya M, Zayed H, Lee S-S. Understanding gene expression and transcriptome profiling of COVID-19: an initiative towards the mapping of protective immunity genes against SARS-CoV-2 infection. Front Immunol. 2021;12:724936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y, Zhao D, Xie WZ, et al. Rap1GAP mediates angiotensin II-Induced cardiomyocyte hypertrophy by inhibiting autophagy and increasing oxidative stress. Oxid Med Cell Longev. 2021;2021:7848027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kundakovic M, Jaric I. The epigenetic link between prenatal adverse environments and neurodevelopmental disorders. Genes. 2017;8(3):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nazzari S, Grumi S, Mambretti F, Villa M, Giorda R, Provenzi L. Maternal and infant NR3C1 and SLC6A4 epigenetic signatures of the COVID-19 pandemic lockdown: when timing matters. Transl Psychiatry. 2022;12:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaiserman AM, Koliada AK. Early-life adversity and long-term neurobehavioral outcomes: epigenome as a bridge? Hum Genomics. 2017;11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosselli M, Keller PJ, Dubey RK. Role of nitric oxide in the biology, physiology and pathophysiology of reproduction. Hum Reprod Update. 1998;4:3-24. [DOI] [PubMed] [Google Scholar]
- 47.Rostamzadeh F, Najafipour H, Yazdani R, Nakhaei S, Alinaghi Langari A. Changes in serum levels of apelin and nitric oxide in hospitalized patients with COVID-19: association with hypertension, diabetes, obesity, and severity of disease. Eur J Med Res. 2022;27:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saeedi Saravi SS, Beer JH. Apelin-potential therapy for COVID-19? J Mol Cell Cardiol. 2020;145:84-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogdanova E, Beresneva O, Galkina O, et al. Myocardial hypertrophy and fibrosis are associated with cardiomyocyte beta-catenin and TRPC6/Calcineurin/NFAT signaling in spontaneously hypertensive rats with 5/6 nephrectomy. Int J Mol Sci. 2021;22:4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waseem M, Shariff MA, Tay ET, et al. Multisystem inflammatory syndrome in children. J Emerg Med. 2022;62:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Ma F, Yokota T, et al. Metabolic reprogramming and epigenetic changes of vital organs in SARS-CoV-2-induced systemic toxicity. JCI Insight. 2021;6:e145027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.SeyedAlinaghi S, Afsahi AM, MohsseniPour M, et al. Late complications of COVID-19; a systematic review of current evidence. Arch Acad Emerg Med. 2021;9:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou J, Wei Y, Zou J, et al. Integrated multi-omics analyses identify key anti-viral host factors and pathways controlling SARS-CoV-2 infection. Res Sq (Preprint). 2022. doi: 10.21203/rs.3.rs-1910932/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oishi Y, Manabe I, Imai Y, et al. Regulatory polymorphism in transcription factor KLF5 at the MEF2 element alters the response to angiotensin II and is associated with human hypertension. FASEB J. 2010;24:1780-1788. [DOI] [PubMed] [Google Scholar]
- 55.Jeong HH, Jia J, Dai Y, Simon LM, Zhao Z. Investigating cellular trajectories in the severity of COVID-19 and their transcriptional programs using machine learning approaches. Genes. 2021;12:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bui LT, Winters NI, Chung M-I, et al. Chronic lung diseases are associated with gene expression programs favoring SARS-CoV-2 entry and severity. Nat Commun. 2021;12:4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein A, Amigo L, Retamal MJ, et al. NPC2 is expressed in human and murine liver and secreted into bile: potential implications for body cholesterol homeostasis. Hepatol. 2006;43:126-133. [DOI] [PubMed] [Google Scholar]
- 59.Prasad K, Ahamad S, Gupta D, Kumar V. Targeting cathepsins: A potential link between COVID-19 and associated neurological manifestations. Heliyon. 2021;7:e08089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huckriede J, de Vries F, Hultström M, et al. Histone H3 cleavage in severe COVID-19 ICU patients. Front Cell Infect Microbiol. 2021;11:694186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ligi D, Giglio RV, Henry BM, et al. What is the impact of circulating histones in COVID-19: a systematic review. Clin Chem Lab Med. 2022;60:1506-1517. [DOI] [PubMed] [Google Scholar]
- 62.Hill RA, Gibbons A, Han U, et al. Maternal SARS-CoV-2 exposure alters infant DNA methylation. Brain Behav Immun Health. 2023;27:100572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sasaki A, Murphy KE, Briollais L, McGowan PO, Matthews SG. DNA methylation profiles in the blood of newborn term infants born to mothers with obesity. PLoS One. 2022;17:e0267946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lorente-Pozo S, Navarrete P, Garzón MJ, et al. DNA methylation analysis to unravel altered genetic pathways underlying early onset and late onset neonatal sepsis. A pilot study. Front Immunol. 2021;12:622599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-gae-10.1177_25168657231184665 for SARS-CoV-2 Covid-19 Infection During Pregnancy and Differential DNA Methylation in Human Cord Blood Cells From Term Neonates by Pedro Urday, Suhita Gayen nee’ Betal, Rochelle Sequeira Gomes, Huda B Al-Kouatly, Kolawole Solarin, Joanna SY Chan, Dongmei Li, Irfan Rahman, Sankar Addya, Rupsa C Boelig and Zubair H Aghai in Epigenetics Insights