-
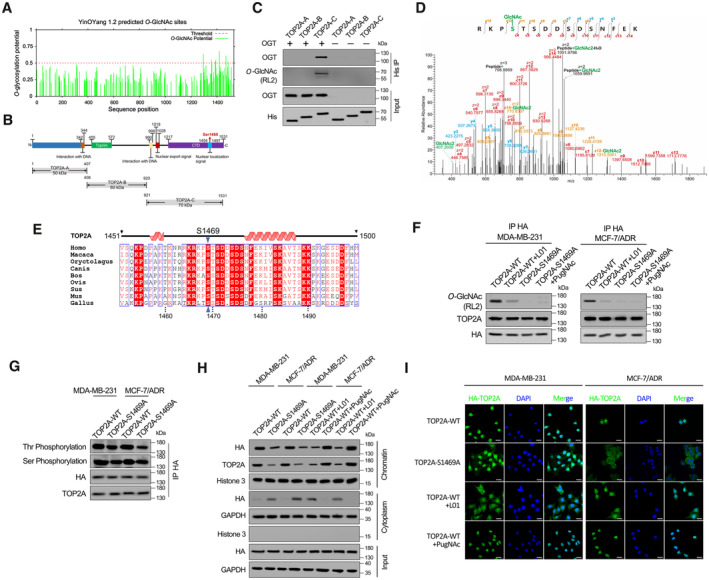
A
O‐GlcNAc sites of human TOP2A predicted using the YinOYang 1.2. The potential O‐GlcNAcylated Ser/Thr residues and the threshold for O‐GlcNAcylation potential were indicated.
-
B
Schematic diagram of key domains in TOP2A. Three truncated variants of TOP2A (TOP2A‐A to C) fused with 6 × His‐tag were generated according to its key domains.
-
C
In vitro glycosylation assay was performed using recombinant His‐tagged truncated variants of TOP2A and purified OGT. The reaction product was detected by co‐immunoprecipitation (co‐IP) and Western blot.
-
D
EThcD‐MS/MS spectrum of the TOP2A CTD peptide with the O‐GlcNAcylation site located on S1469. The matched fragment ions are labeled.
-
E
Alignment of TOP2A CTD sequence among different species.
-
F
MDA‐MB‐231 and MCF‐7/ADR cells which stable transfected with TOP2A‐WT or TOP2A‐S1469A were treated with 50 μM L01 or PugNAc for 48 h. HA‐tag co‐IP was performed, and the immunoprecipitated fractions were analyzed by Western blot for the indicated proteins.
-
G
HA‐tag co‐IP was performed in MDA‐MB‐231 and MCF‐7/ADR cells which stable transfected with TOP2A‐WT or TOP2A‐S1469A. Overall threonine (Thr) and serine (Ser) phosphorylation states of TOP2A were analyzed by Western blot.
-
H
Western blots showing the chromatin binding and subcellular localization of TOP2A‐WT or TOP2A‐S1469A with 50 μM L01 or PugNAc treatment in breast cancer cells.
-
I
Fluorescent staining results showing nuclear localization dependence of TOP2A on O‐GlcNAcylation in MDA‐MB‐231 and MCF‐7/ADR cells. The micrograph scale bar represents 25 μm.