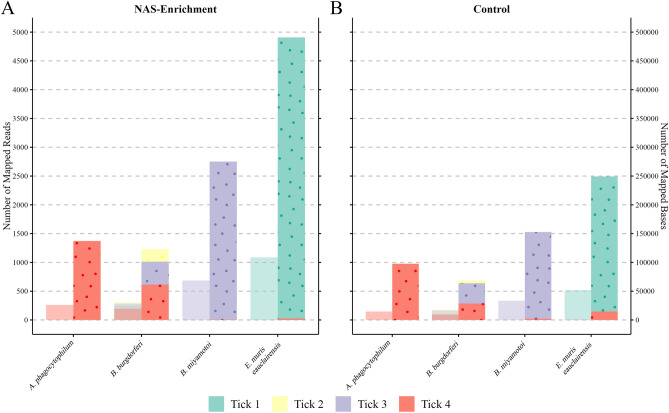
Figure 2.
Enrichment through adaptive sampling yields an increased number of total reads and bases mapping to target tick-borne pathogen taxa. Genomic DNA from individual I. scapularis samples (Ticks 1–4) was sequenced during NAS enrichment and control experiments. (A) Total reads and corresponding bases successfully mapping to each target tick-borne pathogen through NAS enrichment; number of reads are shown in the left columns and total bases shown in right (dotted columns) for each detected tick-borne pathogen genome. (B) Mapped reads (left) and bases (right, dotted columns) generated through unenriched control sequencing. Across each of the four bacterial pathogens detected in these paired NAS and control experiments, adaptive sampling achieved roughly two-fold read and base enrichment in comparison to control sequencing.