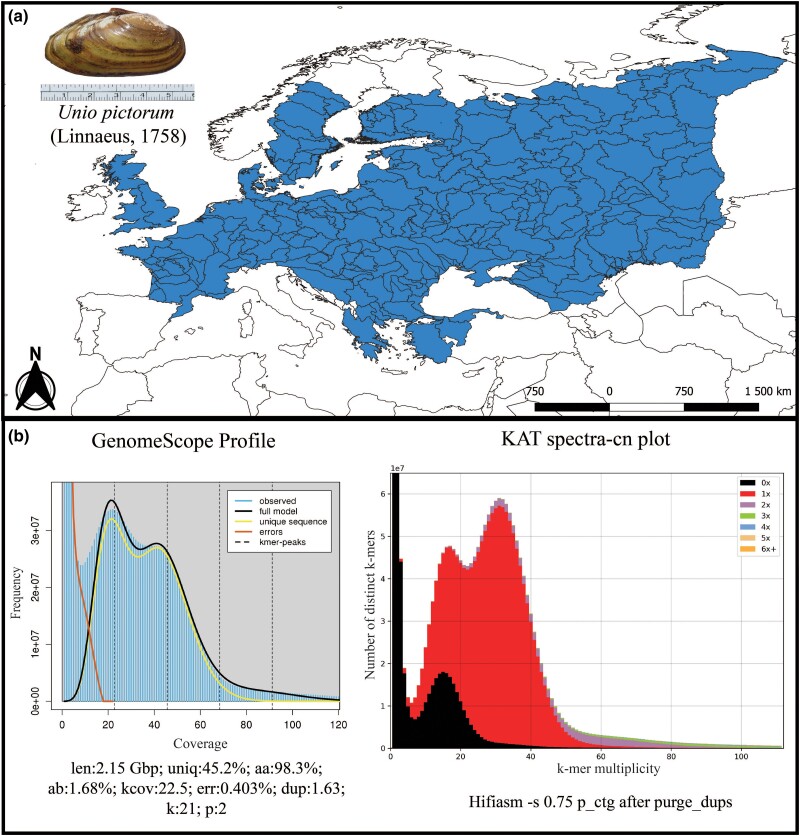
Fig. 1.
A) The map of the potential distribution of Unio pictorum generated by overlapping the points of recent presence records (obtained from 13) with level 5 polygons of the Hydrobasinslayer. On the top left is shown the U. pictorum specimen used for the WGA. B) Left: GenomeScope2 k-mer (21) distribution displaying the estimated genome size (len), homozygosity (aa), heterozygosity (ab), mean k-mer coverage for heterozygous bases (kcov), read error rate (err), the average rate of read duplications (dup), k-mer size used in the run (k:), and ploidy (p:). Right: Assessment of the U. pictorum genome assembly using the KAT comp tool to compare the PacBio Hi-Fi k-mer content within the genome assembly after running purge_dups. Different colors represent the read k-mer frequency in the assembly.