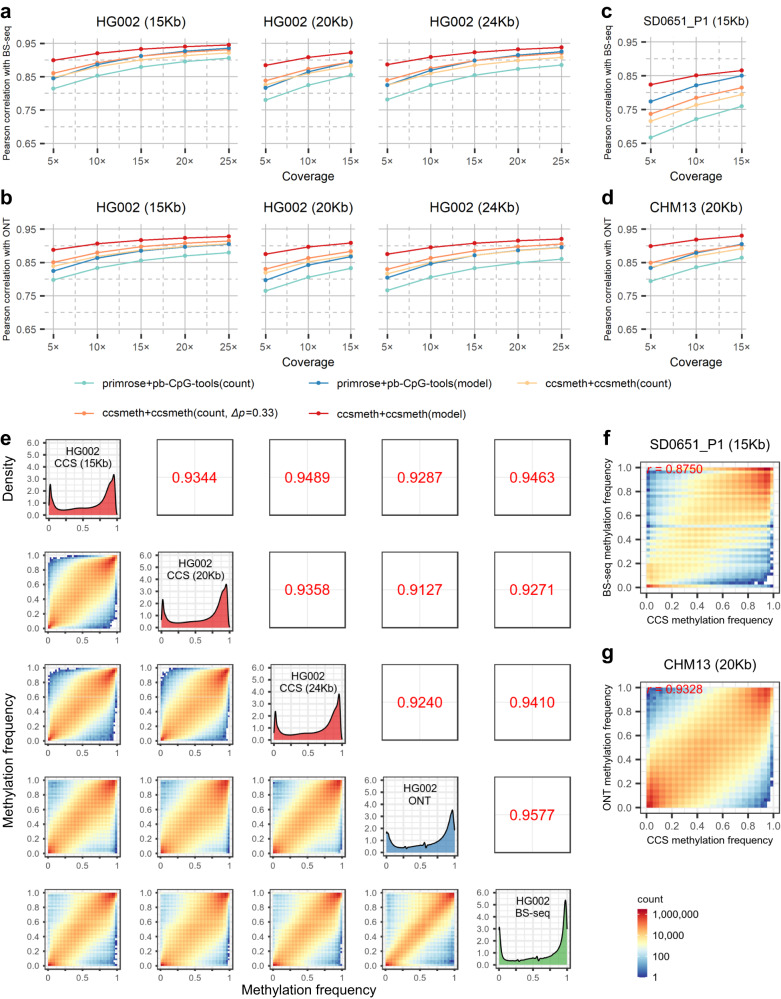
Fig. 3. Evaluation of ccsmeth on 5mCpG detection at genome-wide site level.
a–d Comparing ccsmeth and primrose/pb-CpG-tools against BS-seq and nanopore sequencing under different coverages of HG002, SD0651_P1, and CHM13 CCS reads. ∆p: Difference absolute value between methylated and unmethylated probabilities. Values are the average of 5 repeated tests. The standard deviation values of the multiple repeated tests are in Supplementary Tables 5–12. e Evaluation of ccsmeth model mode against BS-seq and nanopore sequencing using total CCS reads of HG002 (15Kb) (25.6×), HG002 (20 Kb) (17.0×), and HG002 (24 Kb) (28.4×), respectively. Values in upper triangles are Pearson correlations. CCS PacBio CCS sequencing; ONT nanopore sequencing, BS-seq bisulfite sequencing. f Evaluation of ccsmeth model mode against BS-seq using total 19.6× SD0651_P1 (15 Kb) CCS reads. r: Pearson correlation. g Evaluation of ccsmeth model mode against nanopore sequencing using total 16.5× CHM13 (20 Kb) CCS reads. Source data underlying a, b, c, and d are provided as a Source Data file.