Figure 3.
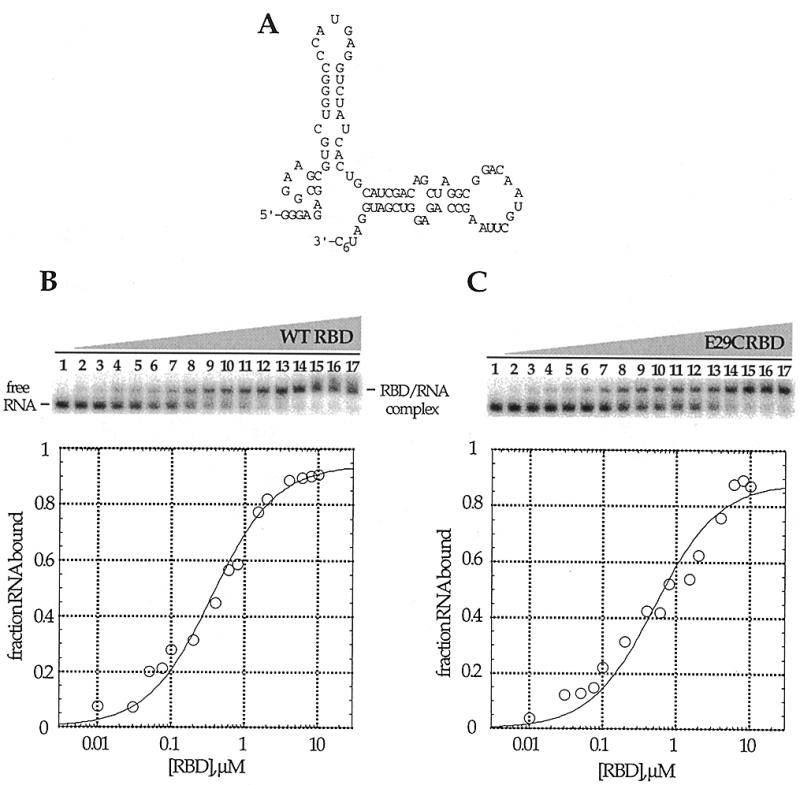
Native gel mobility shifts for the wild-type PKR RBD and PKR RBD (E29C) binding to a 92 nt RNA ligand. (A) Proposed secondary structure of the PKR RNA ligand (8). (B) (Top) Storage phosphor autoradiogram of a representative gel used to separate wild-type PKR RBD-bound from free RNA. Lanes 1–17, 0, 0.01, 0.03, 0.05, 0.075, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 1.5, 2.0, 4.0, 6.0, 8.0 and 10.0 µM wild-type PKR RBD added, respectively. (Bottom) Representative plot of fraction RNA bound by wild-type PKR RBD as a function of protein concentration. The data were fitted to the equation: fraction bound = Θ {[RBD]/([RBD] + Kd)} using the least squares method of KaleidaGraph. (C) (Top) Storage phosphor autoradiogram of a representative gel used to separate PKR RBD (E29C)-bound from free RNA. Lanes 1–17, 0, 0.01, 0.03, 0.05, 0.075, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 1.5, 2.0, 4.0, 6.0, 8.0 and 10.0 µM PKR RBD (E29C) added, respectively. (Bottom) Representative plot of fraction RNA bound by RBD (E29C) as a function of protein concentration.
