Figure 3. Difficulties appraising haplotypes between SVs and neighboring SNVs.
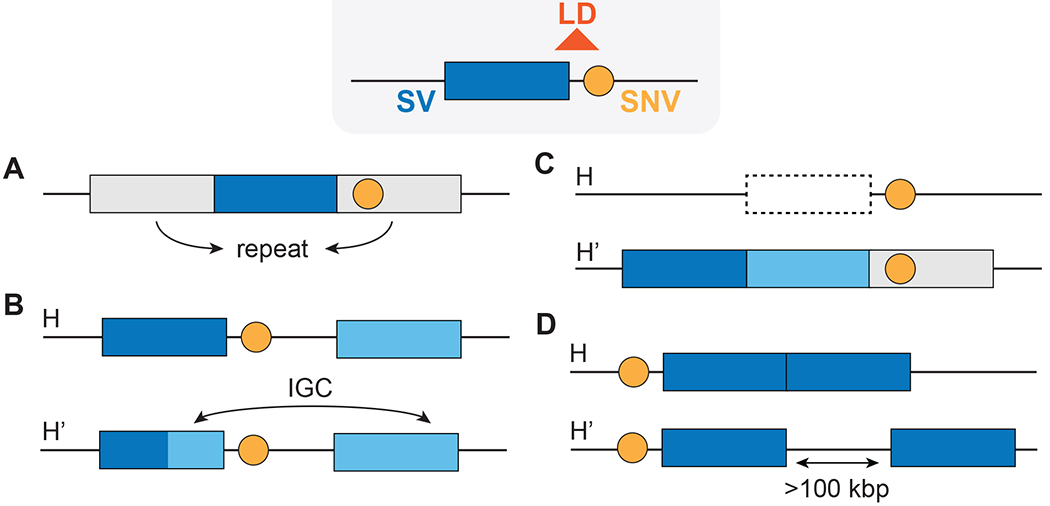
(A) Neighboring SNVs (orange circles) are difficult to detect when an SV (colored rectangle) is embedded in repeat-rich regions. (B, C) Haplotypes can be disrupted by (B) IGC and (C) recurrent deletions (H) and duplication (H’). (D) mCNVs can be in the same locus (H) or several kilobases apart (H’).
