Figure 1. Identification of the putative Hdc enhancers.
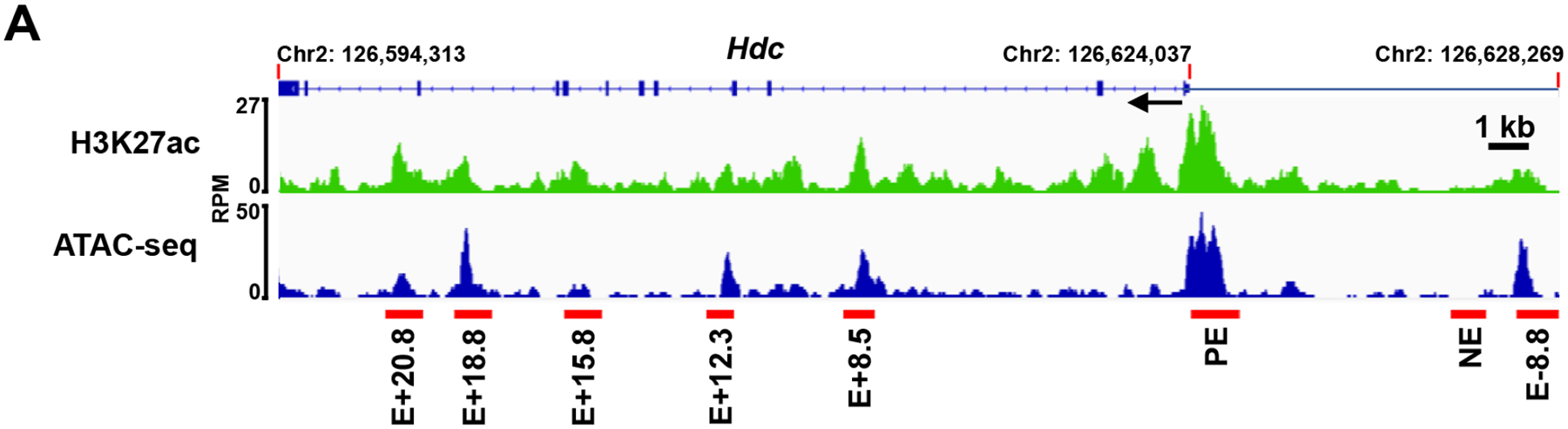
Representative Integrated Genome Viewer (IGV) tracks from H3K27ac CUT&Tag-seq and Omni-ATAC-seq. BMMCs were used for H3K27ac CUT&Tag-seq and Omni-ATAC-seq. Red bars indicate putative Hdc enhancers that show increased H3K27ac modifications and chromatin accessibility. RPM: reads per million mapped reads; E: enhancer; PE: proximal enhancer. The numbers following E indicate the distance (kb) of enhancer to the TSS of the Hdc gene; + means the enhancers are located downstream of the TSS, and − means the enhancers are located upstream of the TSS. One IGV track was from one biological sample, representing two (Omni-ATAC-seq) or three (CUT&Tag-seq) biological samples with similar patterns.
