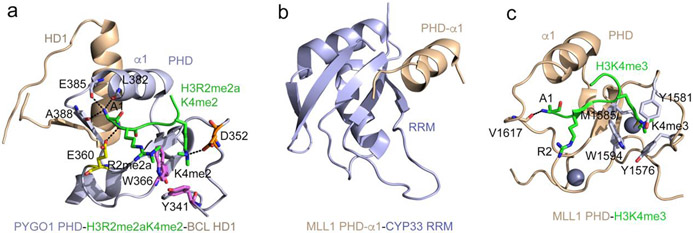
Figure 2: Structural bases of non-histone recognition by PHD fingers.
(a) The ternary complex of the PHD finger of PYGO1 (light blue) with H3R2me2aK4me2 peptide (green) and HD1 (wheat) (PDB ID: 2VPG). Residues of the PHD finger involved in the contact with methylated R2 and K4 of the histone peptide are shown as sticks, labeled and colored magenta, orange and yellow. Hydrogen bonds are indicated by black dash lines. (b) The solution NMR structure of the CYP33 RRM domain (light blue) fused with the α1-helix of the MLL1 PHD3 finger (wheat) (PDB: 2KU7). (c) A ribbon diagram of the structure of the MLL1 PHD finger (wheat) in complex with H3K4me3 peptide (green) (PDB: 3LQJ). The aromatic cage residues are shown as sticks, colored light blue and labeled.