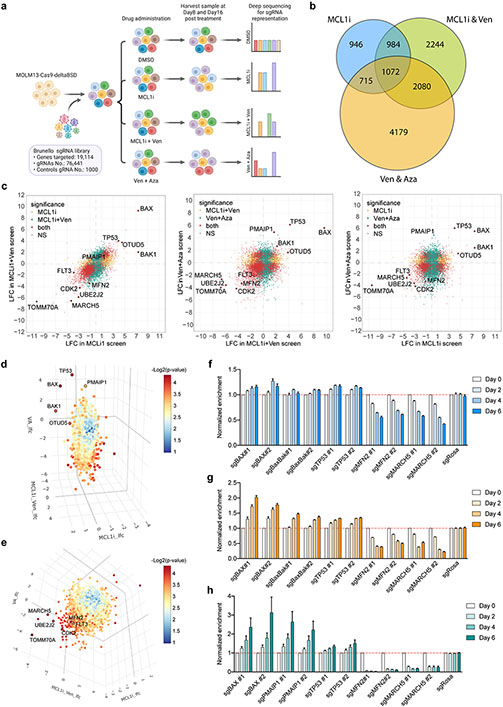
Figure 1. Genome-wide CRISPR/Cas9 loss-of-function screens uncover genes whose ablation synergizes or confers resistance to MCL1i- and Venetoclax- based treatments.
a. Schematic of the screens.
b. Venn diagram depicting the intersection of commonly identified hits in all three screens.
c. Scatter plots showing comparisons of significantly selected genes in MCL1i and MCL1i+Ven screens (left), MCL1i+Ven and Ven+Aza screens (middle), and MCL1i and Ven+Aza screens (right). Genes highlighted in yellow and green are significantly selected in one out of the two indicated screens, while in red are genes significantly selected in both screens shown in each plot. Axes denote the log2 fold change (LFC) of sgRNA representations in drug-treated groups relative to DMSO control. Significance was defined as p < 0.05 in the screen.
d-e. Three-dimensional plots combining the significantly selected hits from all three screens. In panel d, top-ranking positive hits are highlighted, while in e top-scoring negative hits are shown.
f-h. Validation of selected genes using competition-based survival assays in MOLM-13 cells treated with MCL1i (f), MCL1i+Ven (g) or Ven+Aza (h) (n=3, mean ± SD). Normalized enrichment scores were calculated as described in the methods.