Figure 4. Microbial disruption of the classical complement cascade by recruiting inhibitors or degrading complement proteins.
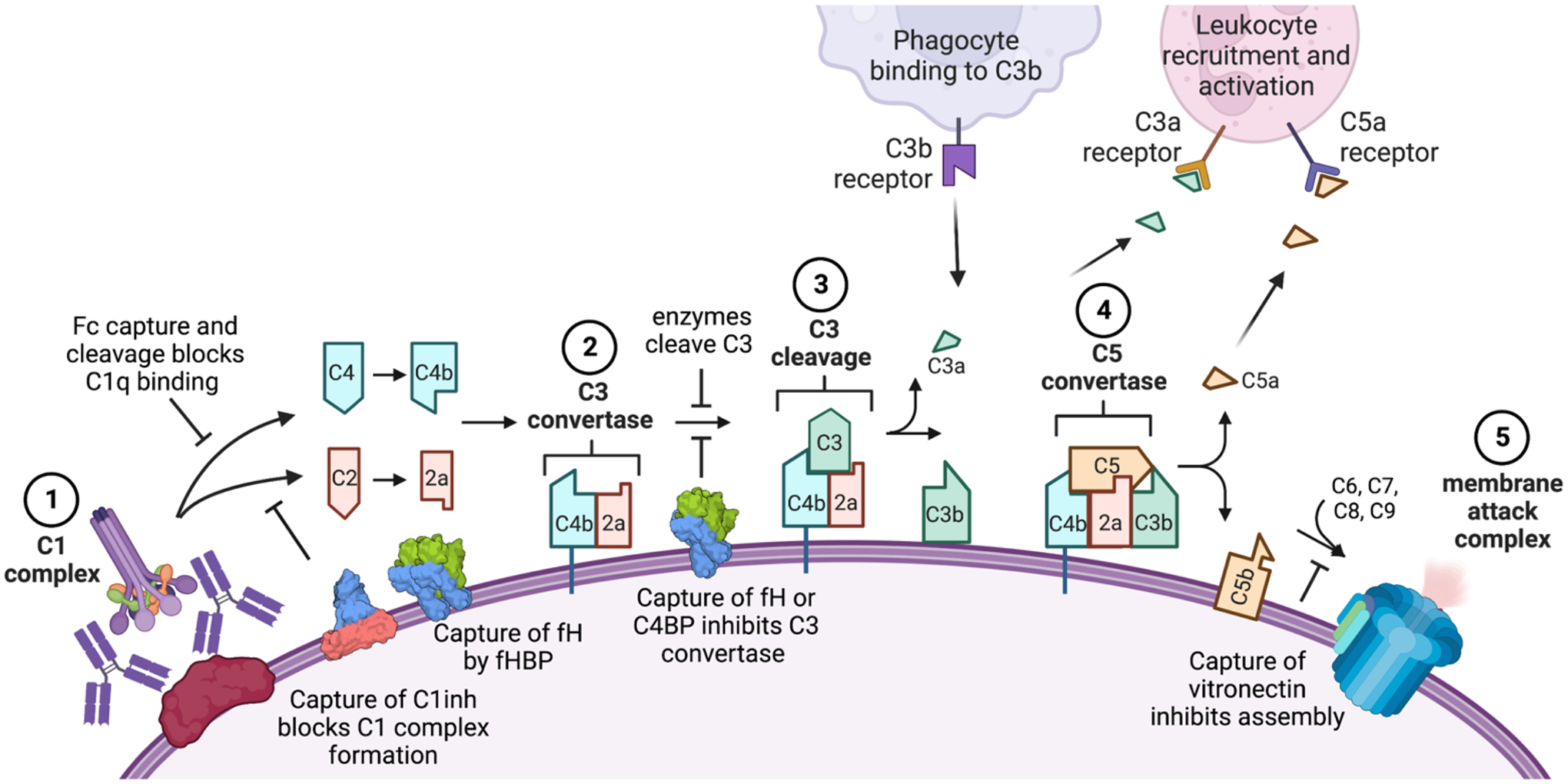
Key steps of the classical pathway of antibody activation shown: (1) the complement proteins C1q and then C1r and C1s bind the microbial surface or IgG/ IgM to form the C1 complex; (2) this cleaves C2 and C4 to produce C4b and C2a which form the C3 convertase; (3) this cleaves C3 to release C3a and deposit C3b covalently on the cell surface; (4) when C3b levels are high, it joins the C3 convertase to form the C5 convertase and deposit C5b on the surface; (5) components C6, C7, C8 and C9 join C5b to form the membrane attack complex and lyse the target cell. The lectin pathway follows a similar cascade but is initiated by the mannose binding lectin complex which recruits C1q, while the alternate pathway results from spontaneous C3 cleavage and C3b deposition to join the cascade at the C3 convertase step using an alternate C3b/Bb complex. Many of these steps can be inhibited by pathogen components, including proteins that bind or cleave the antibody Fc to inhibit C1q recruitment (e.g., protein A, staphylokinase); proteins that recruit host complement regulators (e.g., the Neisseria factor H binding protein [fHBP] which recruits factor H [fH] and B. pertussis Vag8 which recruits C1 inhibitor [C1inh]) and enzymes that degrade complement components (e.g., staphylokinase depletion of C3 away from the microbial surface). Engineering efforts to overcome these strategies include use of Hexabodies whose altered Fc domains favor hexamerization and C1q binding, antibodies altered to resist capture by Fc binding proteins or cleavage by bacterial proteases and antibodies that target microbial evasion proteins to block their functions.
