Figure 4: Nuclear pools of off-chromatin (EZH2-bound) and on chromatin (SF1-bound) β-catenin direct upper zF differentiation in CIMP-high ACC.
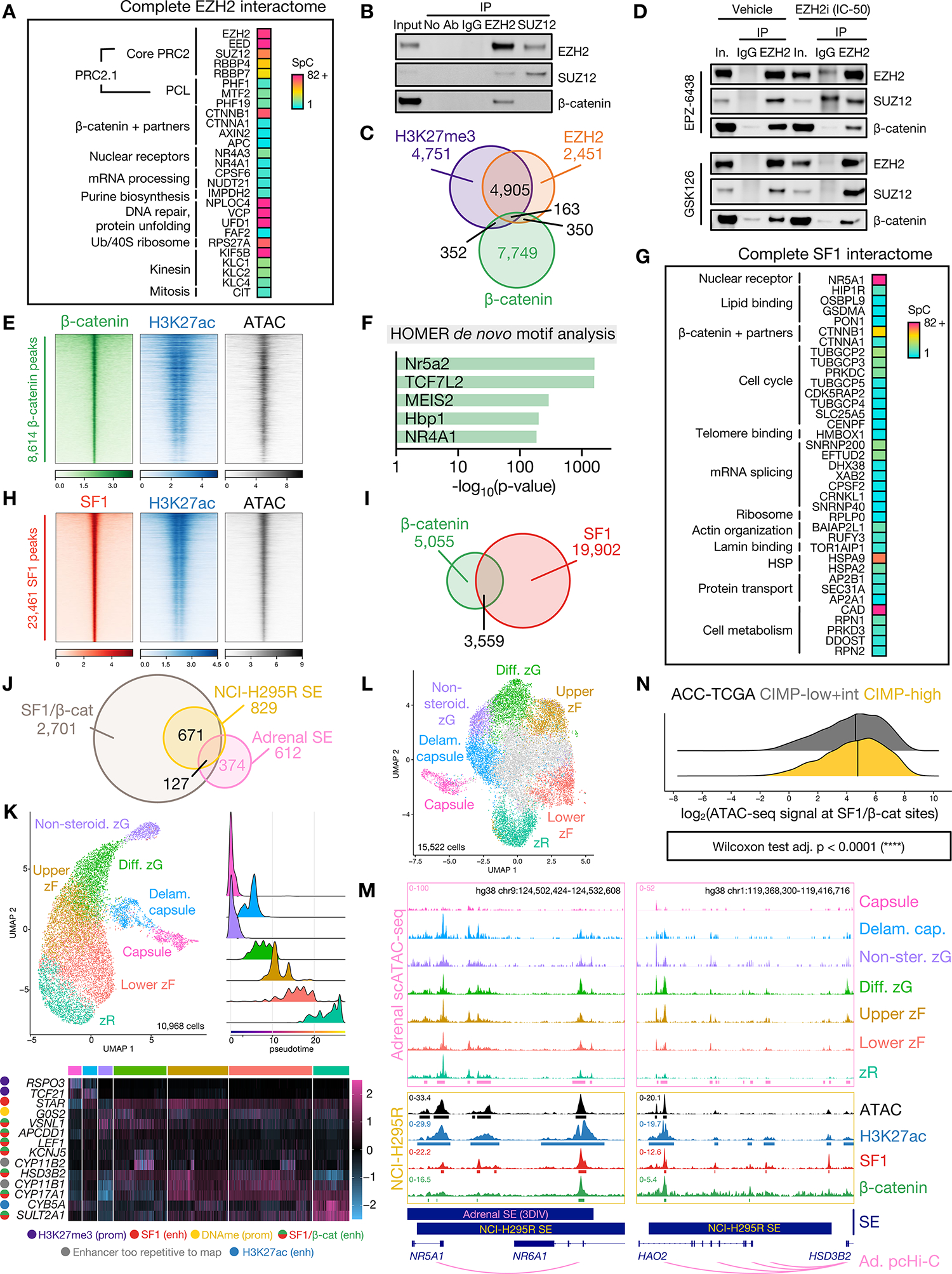
A. Peptides retrieved from EZH2 IP-MS on NCI-H295R nuclear lysates.
B. Representative western blot of NCI-H295R nuclear co-IP, detecting EZH2, SUZ12 and β-catenin. Lanes are 10% input, negative control co-IP (no antibody, IgG), EZH2 IP and SUZ12 IP. n>5 (EZH2 IP), n=2 (SUZ12 IP).
C. Venn diagram of H3K27me3, EZH2, and β-catenin ChIP-seq peaks in baseline NCI-H295R.
D. Representative western blot of EZH2 IP in vehicle- (left) or EZH2i-treated (right, IC-50 dose) NCI-H295R. Lanes are 10% input, negative control IgG IP, EZH2 IP. Higher molecular weight band in EPZ-6438 IgG lane in EZH2/SUZ12 blots is non-specific and emerges when using the same antibody species for IP and western.
E. Heatmap of β-catenin, H3K27ac, and ATAC signal in baseline NCI-H295R at β-catenin peaks, ranked by β-catenin signal. Centered at peak +/− 3 kb window.
F. HOMER motif analysis on baseline NCI-H295R β-catenin peaks.
G. Peptides retrieved from SF1 IP-MS on NCI-H295R nuclear lysates.
H. Heatmap of SF1, H3K27ac and ATAC signal in baseline NCI-H295R at SF1 peaks, ranked by SF1 signal. Centered at peak +/− 3 kb window.
I. Venn diagram of baseline NCI-H295R SF1 and β-catenin peaks.
J. Venn diagram of baseline NCI-H295R SF1/β-catenin peaks, baseline NCI-H295R super-enhancers (SE), and physiologic adrenal SE called by 3DIV on ENCODE samples.
K. Single-cell RNA-seq (scRNA-seq) data from fetal, neonatal and adult human adrenal (31) was integrated and analyzed with comparison to reference markers to identify populations comprising the corticocapsular unit. Top left, scRNA-seq UMAP. Non-steroid.=Non-steroidogenic, Diff.=Differentiated, Delam.=Delaminating. Top right, scRNA-seq pseudotime analysis (origins set for fetal and adult adrenal populations in the capsule and non-steroidogenic zG). Bottom, heatmap of scaled expression of lineage-defining genes across scRNA-seq with epigenetic regulation in NCI-H295R shown left. Prom=promoter, enh=enhancer, DNAme=DNA methylation. Gene regulation by active (H3K27ac only) and SF1- or SF1/β-catenin-bound enhancers was identified by overlap of ChIP-seq with adrenal promoter capture Hi-C (pcHi-C (34)).
L. Single-cell ATAC-seq (scATAC-seq) data from fetal and adult human adrenal (32,33) was integrated and analyzed with comparison to scRNA-seq and reference markers to identify analogous populations comprising the corticocapsular unit. Cells colored in grey in UMAP plot are likely cortical given accessibility within NR5A1 locus though possess ambiguous classification.
M. Example adrenal scATAC-seq and NCI-H295R ATAC, SF1, β-catenin, and H3K27ac tracks across the NR5A1 and HSD3B2 loci in baseline NCI-H295R. Adrenal scATAC peak calls are depicted by pink bars at the bottom of top window, and NCI-H295R peak calls are depicted by bars below each track. 3DIV annotation of adrenal super-enhancers (SE), and NCI-H295R baseline SE are shown by bars below window. Promoter/enhancer contacts from adrenal (Ad.) pcHi-C (34) are depicted below gene annotations.
N. Ridge plot of chromatin accessibility signal at SF1/β-catenin co-targets in ACC-TCGA ATAC-seq samples (n=9, (65)). Line at median.
