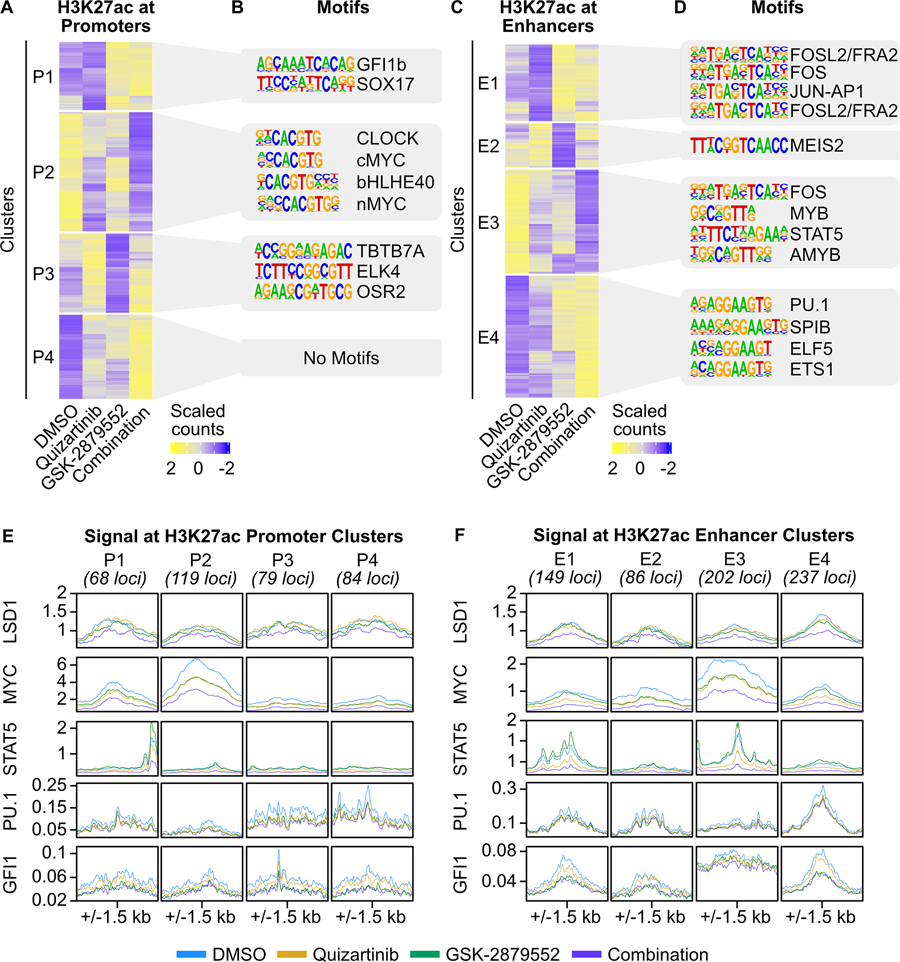
Fig. 2: Discrete components of the response to FLT3/LSD1 inhibition are mediated by promoters and enhancers.
A, MOLM13 cells were treated with quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or an equal volume of DMSO vehicle for 6 hours prior to CUT&Tag for H3K27ac, H3K4me1, and H3K4me3. On the basis of these marks, chromatin was segmented into promoters and enhancers. Unsupervised hierarchical clustering of differential H3K27ac signal at promoters. B, Motif enrichment of promoters with differential H3K27ac signal. Top four de novo motifs with p-value <10−12 are shown. C, D, Same analyses as (A) and (B) were performed at enhancers. E, MOLM13 cells were treated with quizartinib (1 nM), GSK-2879552 (100 nM), the combination, or an equal volume of DMSO for 6 hours. LSD1, MYC, and STAT5 binding was assessed by ChIP-seq. PU.1 and GFI1 binding was assessed by CUT&RUN. Transcription factor binding profiles at promoters with differential H3K27ac identified in (A). F, Transcription factor profiles at enhancers with differential H3K27ac identified in (C).