Figure 1.
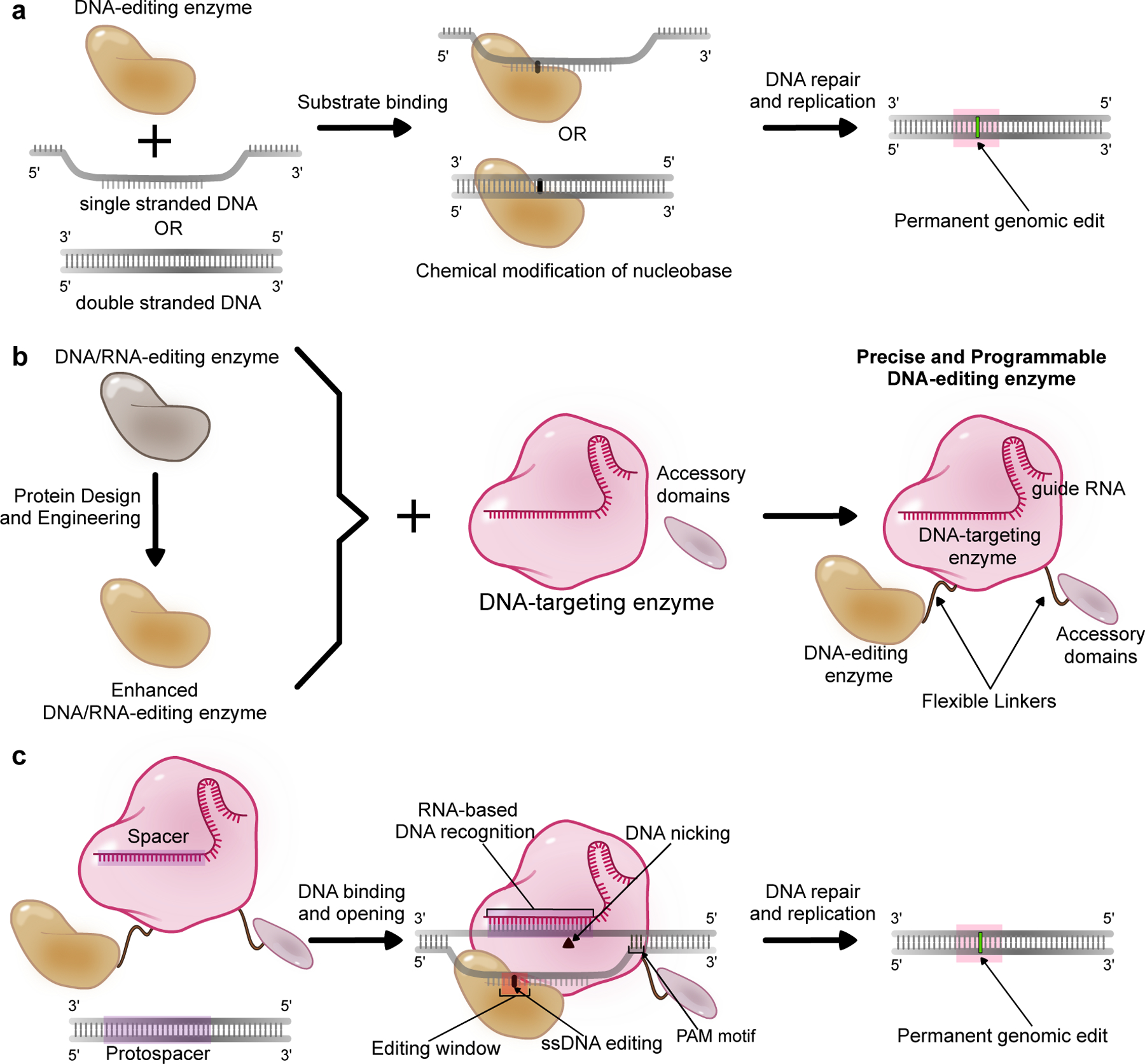
A schematic representation of DNA editing enzymes and their design and application as base editors. (a) “DNA editing enzymes” refer to enzymes that bind to DNA and chemically modify a target nucleobase. The resulting modified nucleobase is processed or interpreted by the cellular repair, replication, or transcriptional machinery as an alternate base, resulting in coding or epigenetic changes in the genome. (b) Naturally-occurring DNA editing enzymes as well as enhanced or engineered variants can be combined with precise and programmable DNA-targeting enzymes such as CRISPR-Cas proteins, along with optional accessory domains (which sometimes are DNA editing enzymes themselves) to produce modular enzyme complexes called base editors. (c) The DNA-targeting module of a base editor (usually CRISPR-Cas9) recognizes and binds to the target genomic locus via base-pairing between a guide RNA (gRNA) molecule and the genomic DNA (the “protospacer”). The protospacer must also be directly next to a protospacer adjacent motif (PAM) to facilitate Cas enzyme binding (for the most commonly used Cas9 system, this PAM sequence is NGG). The Cas protein unwinds the DNA double helix and exposes a small region of single-stranded DNA. If the DNA editing enzyme’s substrate is ssDNA, its DNA editing activity is focused on this ssDNA ’editing window’, and will chemically modify target nucleobases within this window. If, however, the DNA editing enzyme targets double-stranded DNA, it can modify target nucleobases within the general vicinity of the protospacer. Once processed by the cell’s DNA repair and replication machinery, these DNA edits become permanently incorporated into the genome.
