Figure 11.
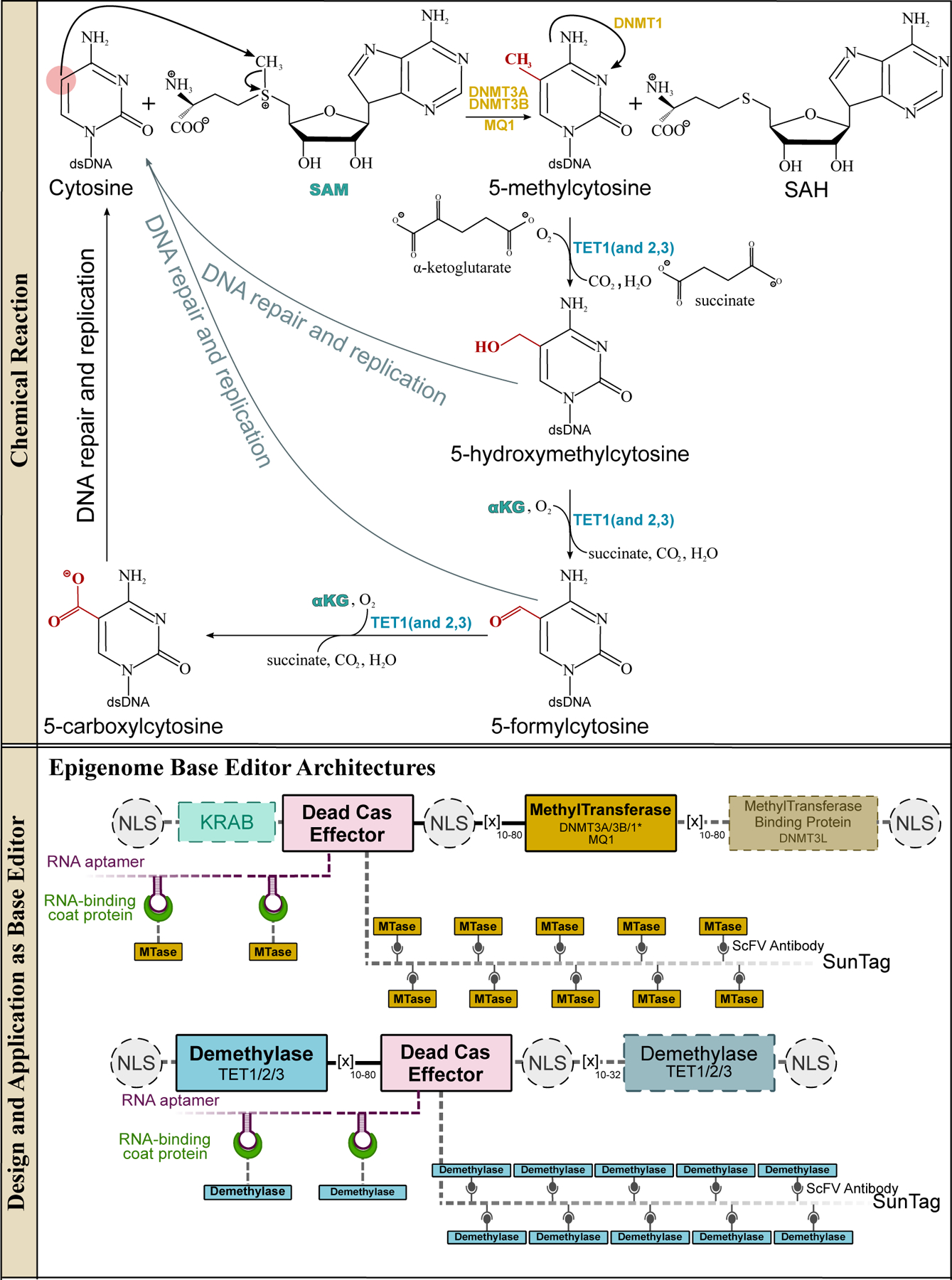
Methyltransferases and methylcytosine dioxygenases and their design and applications in genomic DNA base editing. The methyltransferase enzymes can methylate cytosines in 5′-CpG-3′ islands in dsDNA via a SAM-dependent mechanism to yield 5-methylcytosines (5mC), which act as gene expression suppressors. The demethylation of 5mC is triggered by the TET enzymes, which are iteratively oxygenated to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and finally 5-carboxycytosine (5caC). These oxygenated bases can all be recognized by TDG, which will excise the modified cytosine base. The BER pathway then installs a canonical cytosine base, resulting in demethylation of the 5mC. Overall, these reactions facilitate the full cyclic pathway of C·G→5mC·G and 5mC·G→C·G, which is critical for epigenetic regulation. (Bottom) Representative epigenome base editor architectures are shown, with essential and non-essential components indicated with solid and dashed outlines, respectively.
