Figure 12.
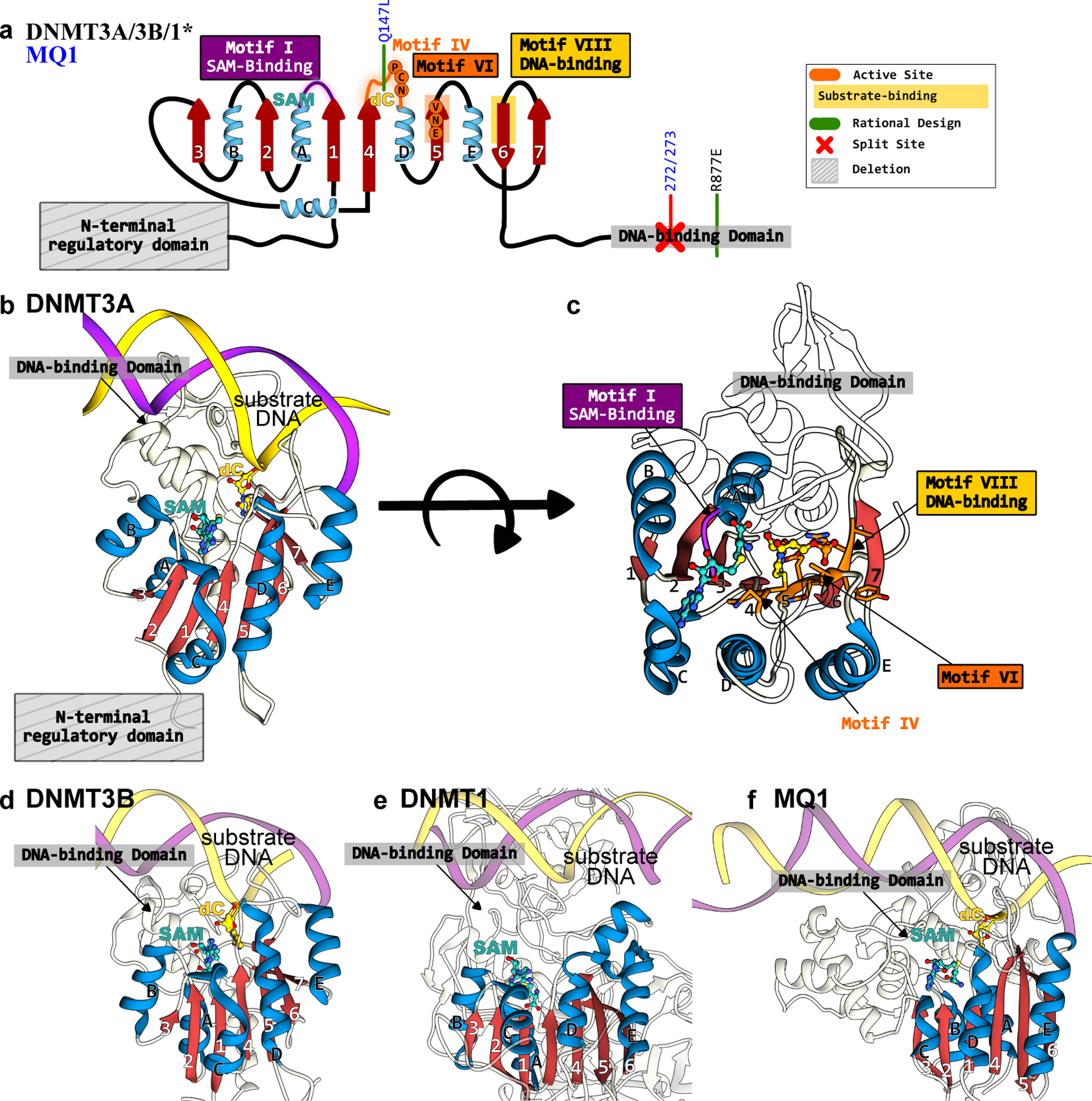
Structures of DNMTs and MQ1 methyltransferases that form the basis of epigenome base editors. (a) The conserved Rossman fold of the DNMT and MQ1 enzymes is color codes as follows: the β-sheet core is in red and peripheral α-helices are in blue. The active site residues, critical DNA- and cofactor-binding sites, and other signature motifs are annotated. Key mutations incorporated to increase activity or specificity are shown in green. (b) Side-view and (c) top-view of DNMT3A correspond to PDB ID: 6BRR(177). (d) Side-view of DNMT3B structure(6U8P(178)). (e) Side-view of DNMT1 structure (3PTA(179)). (f) Side-view of MQ1 structure, generated using Alphafold2(37) with the substrate dsDNA modeled in using its structural homolog(177).
