Figure 13.
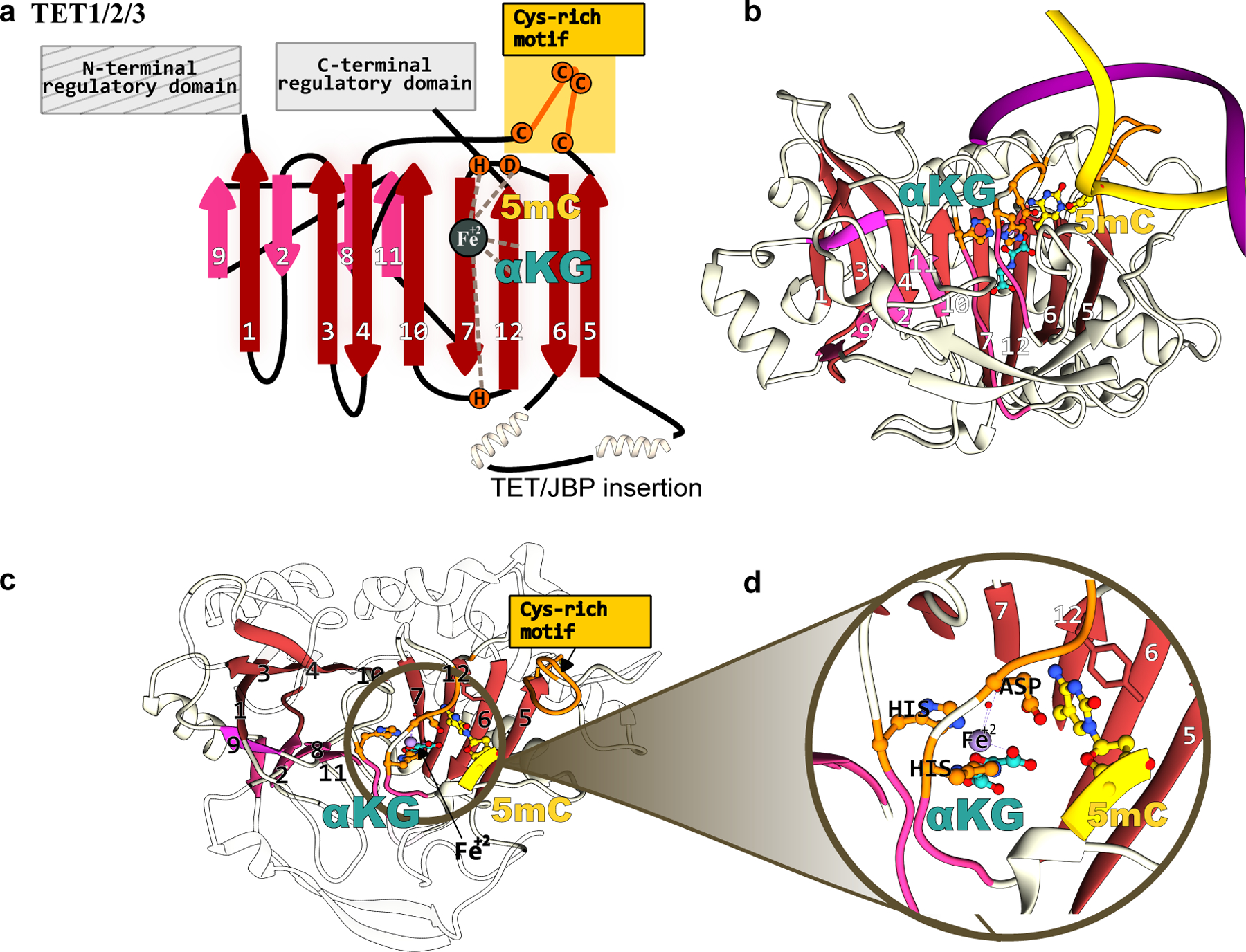
Structure of TET methylcytosine dioxygenases that form the basis of epigenome base editors. (a) The conserved jelly-roll fold of the TET1 enzyme consists of a β-sheet core, shown in red and pink. The active site residues, critical DNA- and cofactor-binding sites, and other signature motifs are annotated. (b) Side-view, (b) top-view, and (c) zoomed-in structural model of the TET1-DNA complex. The TET1 structure was predicted using Alphafold2(37) and the substrate dsDNA was modeled in using its structural homolog(180).
