Figure 3.
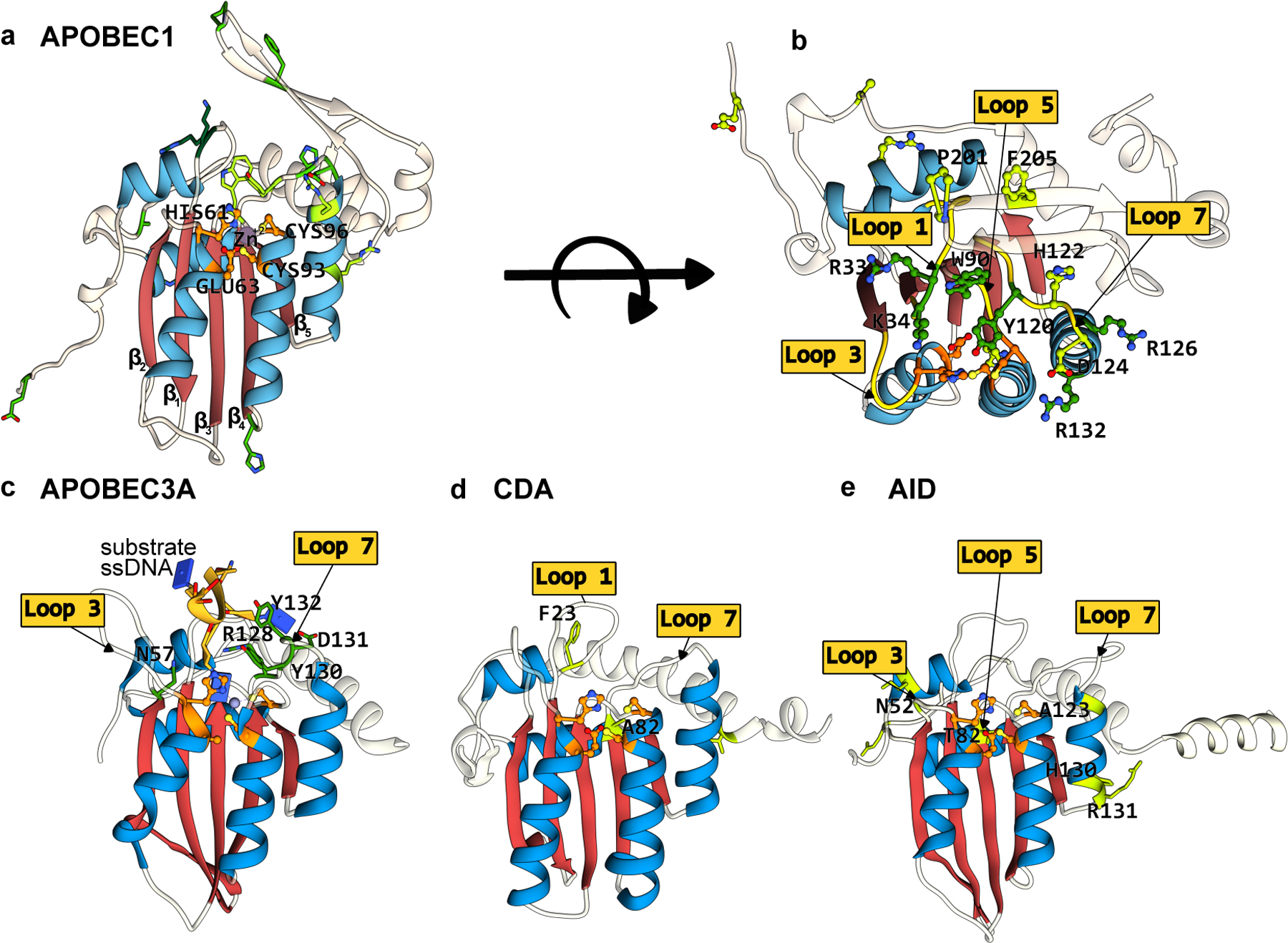
Structures of APOBEC/AID deaminases, which form the basis of the most extensively used CBEs. The conserved CDA fold is color-coded as follows: the β-sheet core is in red and the peripheral α-helices are in blue. The active site residues are shown in orange, with mutations that have enhanced certain properties of these deaminases (when used as a base editor) shown in green. (a) Side-view of the APOBEC1 structure (generated using Alphafold2(37)) depicting the CDA-family fold and the active site residues. (b) Top-view of the APOBEC1 structure, highlighting key amino acid mutations shown to enhance its base editing activity and the critical active site loops where these residues are located. (c-e) Side-view of APOBEC3A-ssDNA(PDB ID:5KEG(60)), CDA, and AID (generated using Alphafold2(37)), respectively. Active site loops and key amino acids are highlighted. Labels for active site residues are omitted for clarity. (cf. Figure 2)
