Figure 5.
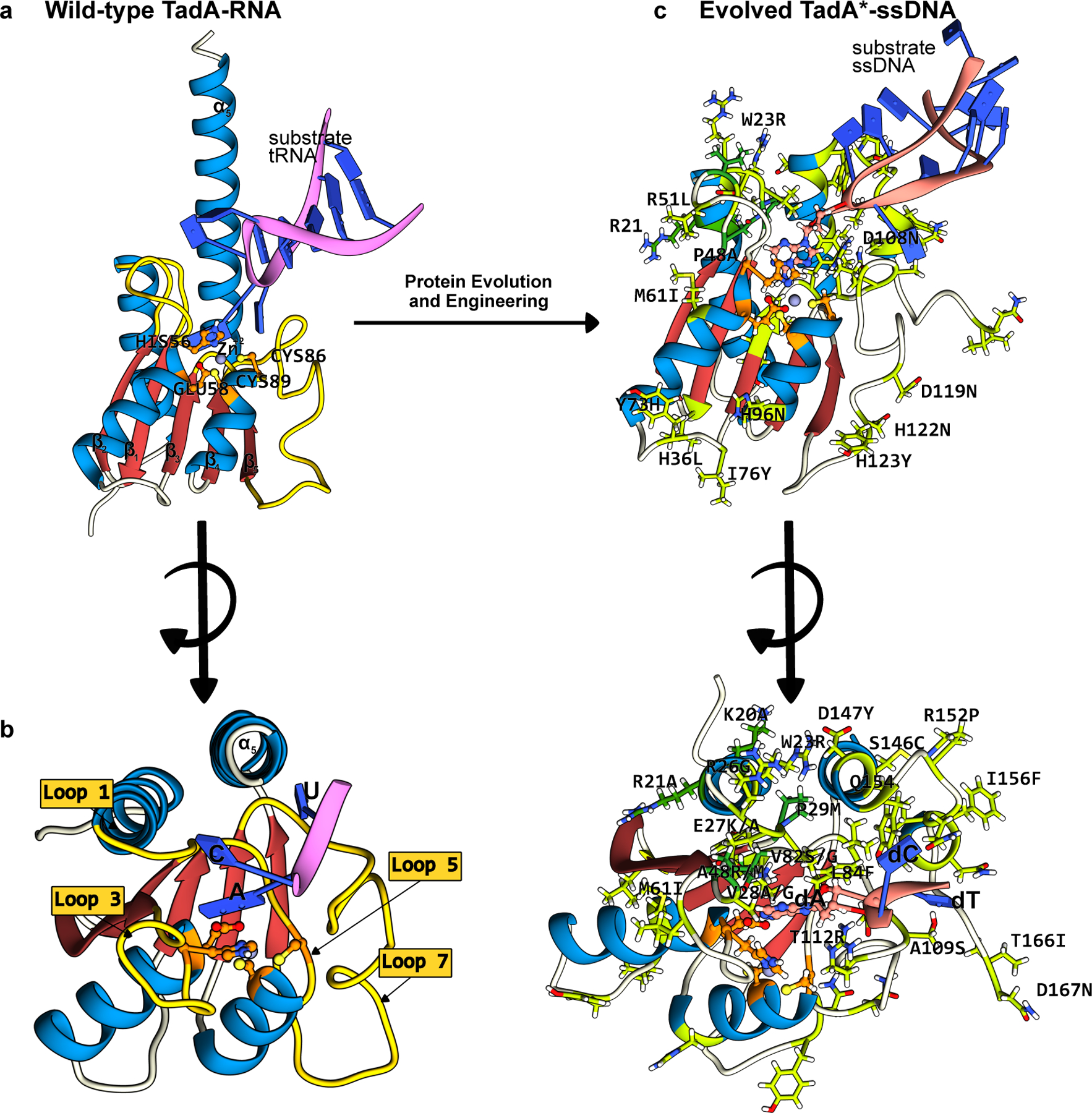
Structures of the wt-ecTadA enzyme bound to its substrate RNA, and the evolved TadA* bound to an ssDNA substrate. The conserved CDA fold is color-coded as follows: the β-sheet core is red and the peripheral α-helices are blue. The active site residues are shown in orange, with the mutations that have enhanced certain properties of the deaminase (when used as a base editor) shown in green. (a) Side-view of the wt-TadA-tRNA complex, generated using a combination of Alphafold2 predictions(37) and PDB ID:2B3J(82). (b) Top-view of the wt-TadA-tRNA complex. The RNA structure beyond the bases in the active site is omitted for clarity. (c) Side-view of the TadA*-ssDNA complex, generated from PDB ID:6VPC(75). (d) Top-view of the TadA*-ssDNA complex. The DNA structure beyond the bases in the active site is omitted for clarity. (cf. Figure 4)
