Fig. 1.
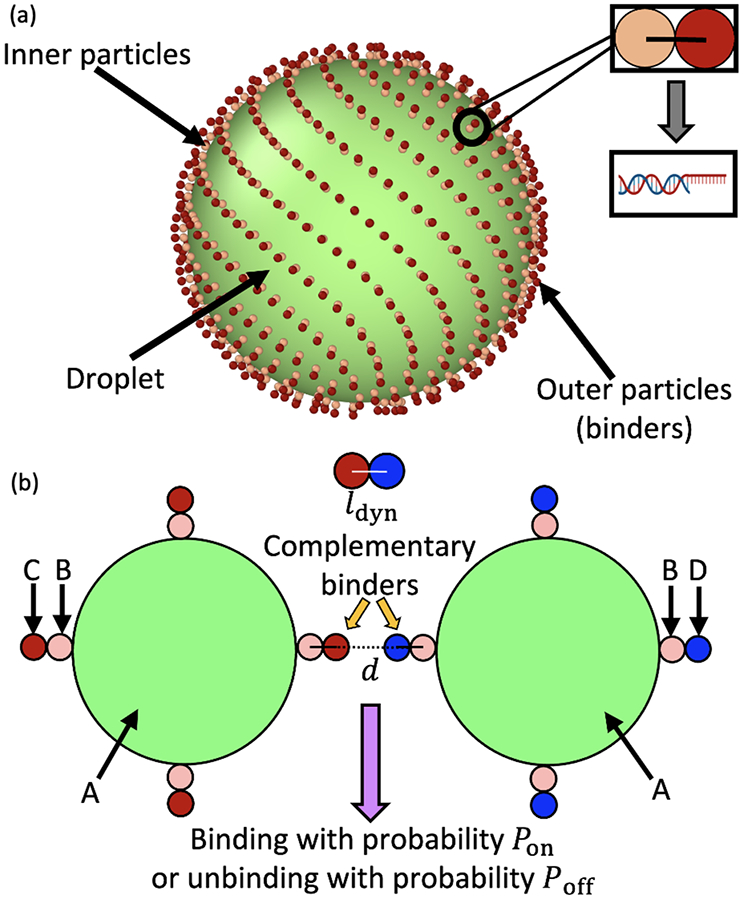
(a) The initial configuration of a droplet with binders adhered to its surface, arrayed in their initial “Fibonacci” structure. As shown in the inset, each binder consists of two constituent particles, which in the case of DNA corresponds to a spacer double-stranded sequence and a single stranded sequence which is available to bind to a complementary strand. (b) A schematic showing dynamic binding between the outer binder particles of two droplets. Different particle types used in our python framework are schematically labeled A–D, with C and D representing a pair of complementary binders.
