Figure 4.
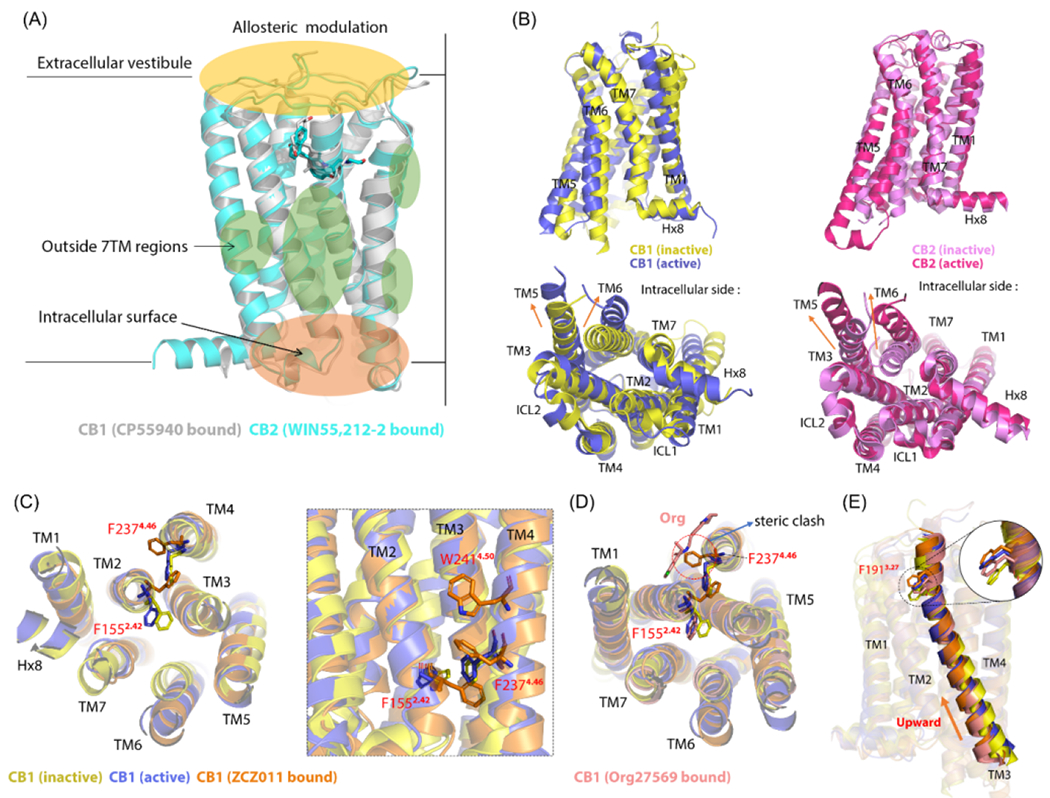
Molecular mechanism for cannabinoid receptor (CBR) allostery: 1. (a) CBR allosteric modulators (AMs) mainly bind to three regions: extracellular vestibule, outside transmembrane (TM) 7 regions, and intracellular surface. Cryo-electron microscopy (cryo-EM) overlapped structures of CP55940-bound CB1R [in grey; Protein Data Bank (PDB): 7wv9] and WIN55,212–2–bound CB2R (in cyan; PDB: 6pt0). (b) TM6 outward movement and TM5 arrangement in overlapped CB1R and CB2R in the active and inactive state. The crystal structures of CB1R in inactive state are shown in yellow (PDB: 5tyz), whereas the active state is shown in blue (PDB: 5xr8). The crystal structures of CB2R in the inactive state are shown in violet (PDB: 5zty), whereas the active state is shown in pink (PDB: 6pt0). Both CB1R and CB2R have significant outward movement of TM6. For the TM5 arrangement, an elongation occurs in CB1R, whereas an outward movement occurs in CB2R. (c) TM2 arrangement in ZCZ011-bound CB1R (in orange; PDB: 7wv9) compared with inactive and active CB1R with the reorientation of F1552.42 and F2374.46. The left panel shows the intracellular view, and the right panel shows the same residue positions from the view of the TM domain. W2414.50 forms an aromatic stack with F2374.46. (d) Org27569-bound CB1R (in salmon; PDB: 6kqi) causes a steric clash with F2374.46, thus influencing F1552.42 reorientation and impeding the TM2 arrangement. (e) Upward movement of TM3 in four overlapped CB1Rs (F1913.27 as reference). The sequence from top to bottom is: ZCZ011-bound CB1R, active CB1R, Org–bound CB1R, and inactive CB1R. Figure based on 15,21,23.
