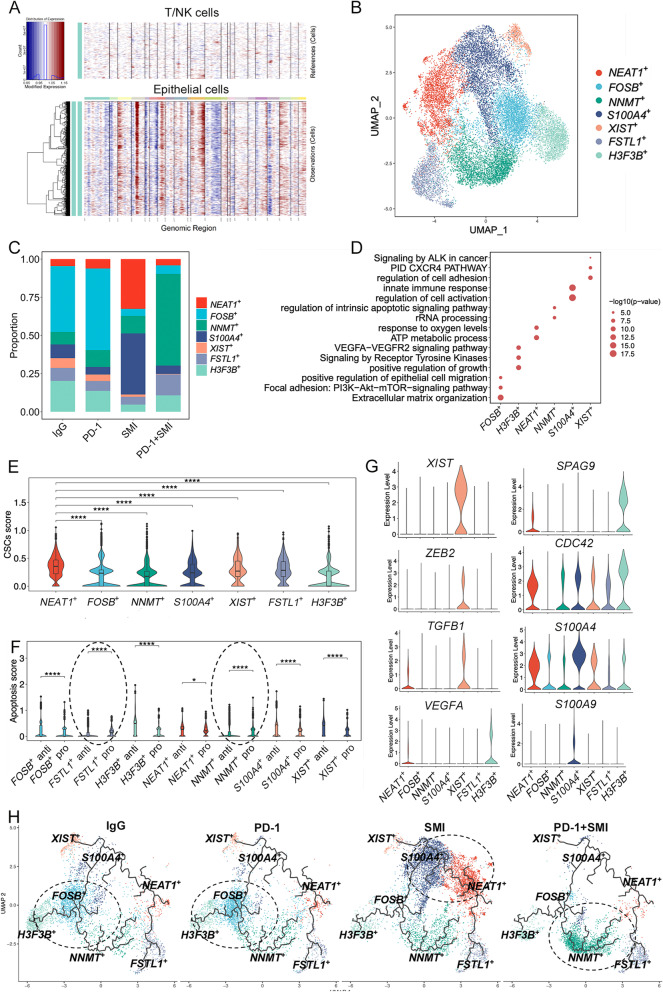
Fig. 4.
Seven subclusters of malignant epithelial cells were identified among different treatments. (A) Malignant epithelial cells were distinguished with T/NK cells by inferred large-scale chromosomal copy number variations (CNVs). (B) UMAP of the seven malignant epithelial clusters: NEAT1+ tumor cells (NEAT1+), FOSB+ tumor cells (FOSB+), NNMT+ tumor cells (NNMT+), S100A4+ tumor cells (S100A4+), XIST+ tumor cells (XIST+), FSTL1+ tumor cells (FSTL1+), H3F3B+ tumor cells (H3F3B+). (C) Relative proportion of seven malignant epithelial clusters in four different treatments. IgG: immunoglobulin G isotype control; PD-1: PD-1 immune-checkpoint blockade antibody; SMI: SMI monotherapy; PD-1+SMI: combination of anti-PD-1 and SMI. (D) Dot plot showing the top representative biological pathways of seven malignant epithelial clusters based on DEGs using Metascape analysis. (E) Violin plot showing the highest cancer stem cells (CSCs) score in NEAT1+ tumor cells compared with that in other subclusters. ****P < 0.0001. (F) Violin plot showing the mean score of the apoptosis or anti-apoptosis signature across seven malignant epithelial clusters. NNMT+ and FSTL1+ tumor cells displayed significantly higher pro-apoptosis scores than anti-apoptosis score, while other subcluster cells displayed significantly higher anti-apoptosis scores than pro-apoptosis scores. *P < 0.05, ****P < 0.0001. (G) Violin plot showing expression of XIST, ZEB2, TGFB1, VEGFA, SPAG9, CDC42, S100A9, and S100A4 in seven malignant epithelial clusters. (H) Unsupervised transcriptional trajectory of seven malignant epithelial clusters predicted by Monocle 3 in four different treatments