Abstract
Background and Aims
Grasses of the Festuca genus have complex phylogenetic relations due to morphological similarities among species and interspecific hybridization processes. Within Patagonian fescues, information concerning phylogenetic relationships is very scarce. In Festuca pallescens, a widely distributed species, the high phenotypic variability and the occurrence of interspecific hybridization preclude a clear identification of the populations. Given the relevance of natural rangelands for livestock production and their high degradation due to climate change, conservation actions are needed and knowledge about genetic variation is required.
Methods
To unravel the intraspecific phylogenetic relations and to detect genetic differences, we studied 21 populations of the species along its natural geographical distribution by coupling both molecular [internal transcribed spacer (ITS) and trnL-F markers] and morpho-anatomical analyses. Bayesian inference, maximum likelihood and maximum parsimony methods were applied to assemble a phylogenetic tree, including other native species. The morphological data set was analysed by discriminant and cluster analyses.
Key Results
The combined information of the Bayesian tree (ITS marker), the geographical distribution of haplotype variants (trnL-F marker) and the morpho-anatomical traits, distinguished populations located at the margins of the distribution. Some of the variants detected were shared with other sympatric species of fescues.
Conclusions
These results suggest the occurrence of hybridization processes between species of the genus at peripheral sites characterized by suboptimal conditions, which might be key to the survival of these populations.
Keywords: Intraspecific genetic differentiation, phylogenetic relationships, ITS, chloroplast DNA markers, morpho-anatomical analyses, plant traits, Festuca pallescens, Festuca, Patagonia, glacial refugia, hybridization, speciation
INTRODUCTION
Dryland ecosystems cover almost half of Earth’s land surface and house nearly 40 % of humanity, with outstanding importance for socioeconomic activities (Bradford et al., 2020; Maestre, 2021). Natural rangelands that develop in dryland areas constitute most of the world’s grasslands and are degraded due to overexploitation (Grau and Aide, 2008; James et al., 2013). In addition, plant communities of dryland ecosystems are highly sensitive to changes in temperature and precipitation regimes, such as those occurring due to global climate change (Magrin et al., 2014; Golodets et al., 2015). Increasing the knowledge of evolutionary processes in populations of key grassland species is necessary to both help predict upcoming changes and contribute to solutions.
Broadly distributed grassland species, covering environmental gradients and heterogeneous habitats, provide an interesting experimental design to study evolution and adaptation to suboptimal conditions (Maestre et al., 2012). Within these habitats, species have adapted in unique ways by developing deep and efficient root systems and water storage structures, or by going dormant during dry periods, to evade, avoid or resist drought (Davies et al., 2012; Moreno and Bertiller, 2015). These species tend to present large phenotypic and genetic variation (Manel et al., 2012). The level of genetic differentiation among central populations of the same species tends to be low due to historical persistence and the constant genetic exchange between individuals, but habitat heterogeneity could result in genetically distinct ecotypes (Hufford and Mazer, 2003; Eckert et al., 2008; Jakob et al., 2009). The continuous stress that populations undergo in dryland ecosystems may lead to adaptive changes and eventually even speciation processes (Hoffmann and Hercus, 2000). Natural hybridization may be a mechanism to promote adaptation, particularly in small and isolated populations that develop near the limits of their distribution range (Thompson et al., 2010). Specifically, glacial refugia during the Quaternary could have developed the conditions for the occurrence of ancient hybridization processes (Liu et al., 2018; Shepherd et al., 2022). Ultimately, hybridization may lead to adaptive evolution and speciation, which allows the new species to colonize extreme habitats to which neither of the parental species is adapted (Rieseberg et al., 2003; Liu et al., 2011). The recognition of ecotypes or hybrids can be complex, but the use of molecular markers complemented by the analysis of morphological character expression permits the reconstruction of phylogenetic trees (Pourebrahimi et al., 2022), thus making it possible to assign taxonomic status.
The Patagonian region constitutes the largest arid or semiarid ecosystem in southern South America. Climatic gradients mainly determined by the Andean mountains and a complex geomorphology give rise to a variety of environments and soil and vegetation types (Villalba et al., 2003; Gaitán et al., 2020). Plant communities in Patagonia are represented by two main functional groups: shrubs and perennial grasses (Bertiller and Bisigato, 1998). Natural rangelands dominated by tussock grasses and scattered shrubs are the main input for cattle raising and have been intensively used since the early 1900s (Defossé et al., 1990). Due to overgrazing and the effects of climate change, these valuable habitats undergo degradation and desertification, meaning that key species of grasses become less abundant due to a drastic reduction of their populations (Gaitán et al., 2017; Oliva et al., 2020). As perennial dominant grasses lose habitat, grasslands deteriorate and different species, particularly shrubs, may establish and dominate (Gonzalez and Ghermandi, 2021).
Festuca pallescens is a dominant allohexaploid species from natural rangelands of Patagonia (Dubcovsky and Martínez, 1992). This keystone species has a wide distribution and inhabits many diverse environments. It has been well studied and described morphologically and physiologically (Somlo et al., 1985; Bertiller et al., 1990; Fernández et al., 2004, 2006; Caballé et al., 2011; López et al., 2019, 2021), but information about its genetic variability is scarce (Dubcovsky and Martínez, 1991, 1992; López et al., 2020). Phenotypic variation among populations analysed in a common garden was observed and the molecular identification of a hybrid ecotype was carried out (López et al., 2018, 2020). This ecotype is found in a peripheral population clustered with another closely related polyploid, Festuca argentina, a sympatric species of F. pallescens (López et al., 2018). However, this previous study covered only part of the distribution range. Here, by extending the analysis to the whole natural range, we aim to analyse intraspecific variation and to reconstruct interspecific phylogenies involving other native species. Our main hypothesis is that the wide distribution range of this species, covering highly heterogeneous habitats, promoted genetic differentiation among populations. We also hypothesize that hybridization with sympatric species is possible in peripheral areas. We combined a molecular study of populations encompassing the whole distribution range with a thorough morphological analysis to describe intraspecific variation.
MATERIALS AND METHODS
Sampling strategy
Twenty-one populations of Festuca pallescens covering the distribution range of the species were analysed (Table 1, Fig. 1). Leaves were collected for DNA extraction and sequencing. Sampled individuals were identified in the field based on exomorphological characters described for the species (Nicora, 1978; Catalán and Müller, 2012). Additionally, four individuals of the sympatric hexaploid species Festuca gracillima, from the southernmost area of F. pallescens’ distribution, were collected and included in the analysis.
Table 1.
Geographic locations of the sampling sites of Festuca pallescens populations. GenBank accession numbers are included.
| Population | Abbreviation | Sample site | Latitude (south) | Longitude (west) | GenBank accession no. | |
|---|---|---|---|---|---|---|
| ITS | trnL-F | |||||
| 1 | ACA* | Aguas Calientes, Neuquén, Argentina | 36°41ʹ2″ | 70°36ʹ37″ | OP081817 | KX701976 |
| 2 | LMI | Los Miches, Neuquén, Argentina | 37°6ʹ36″ | 70°49ʹ3″ | OP081817 | KX701976 |
| 3 | HUI* | Huinganco, Neuquén, Argentina | 37°12ʹ17″ | 70°37ʹ12″ | OP081817 | KX701976 |
| 4 | CHO | Chos Malal, Neuquén, Argentina | 37°21ʹ38″ | 70°7ʹ23″ | OP081817 | KX701976 |
| 5 | CLI* | Catan Lil 2, Neuquén, Argentina | 39°45ʹ40″ | 70°37ʹ7″ | KX688222 | KX701976 |
| OP081824 | ||||||
| 6 | SRA | San Ramón Ranch, Río Negro, Argentina | 41°10ʹ32″ | 70°59ʹ1″ | KX688222 | KX701976 |
| 7 | PIL* | Pilcaniyeu Experimental Field, Río Negro, Argentina | 41°3ʹ40″ | 70°31ʹ1″ | KX688222 | KX701976 |
| 8 | MON | Montoso, Chubut, Argentina | 42°42ʹ52″ | 71°01ʹ59″ | KX688222 | KX701976 |
| 9 | JAC | Ingeniero Jacobacci, Río Negro, Argentina | 41°55ʹ9″ | 69°12ʹ58″ | KX688222 | KX701976 |
| KX701978 | ||||||
| 10 | SOM | Somuncura Plateau, Río Negro, Argentina | 41°25ʹ1″ | 66°58ʹ1″ | KX688222 | KX701976 |
| 11 | YAG | Yague, Chubut, Argentina | 42°57ʹ0″ | 71°12ʹ0″ | KX688222 | KX701976 |
| OP081824 | ||||||
| 12 | APE* | Arroyo Pescado, Chubut, Argentina | 43°2ʹ49″ | 70°58ʹ2″ | KX688222 | KX701976 |
| 13 | CRO* | Cronómetro, Chubut, Argentina | 43°14ʹ19″ | 71°4ʹ54″ | OP081820 | KX701976 |
| 14 | GOC | Gobernador Costa, Chubut, Argentina | 44°1ʹ12″ | 70°51ʹ36″ | KX688222 | KX701976 |
| 15 | RPI | Río Pico, Chubut, Argentina | 44°8ʹ24″ | 71°26ʹ24″ | KX688222 | KX701976 |
| 16 | FON* | Fontana, Chubut, Argentina | 44°56ʹ46″ | 71°31ʹ09″ | OP081819 | KX701976 |
| 17 | RMA* | Río Mayo, Chubut, Argentina | 45°28ʹ12″ | 69°49ʹ48″ | KX688222 | KX701976 |
| OP081816 | OP081821 | |||||
| OP081822 | ||||||
| OP081824 | ||||||
| 18 | LBA* | Mte Lago Bs As, Santa Cruz, Argentina | 46°36ʹ0″ | 71°7ʹ48″ | KX688222 | KX701976 |
| 19 | LCO | Laguna Colorada, Santa Cruz, Argentina | 51°40ʹ15″ | 69°53ʹ58″ | KX688222 | KX701976 |
| 20 | BLE* | Bajo la Leona, Santa Cruz, Argentina | 51°31ʹ48″ | 69°42ʹ4″ | KX688222 | OP081823 |
| 21 | GAI* | Guer Aike, Santa Cruz, Argentina | 51°37ʹ33″ | 69°37ʹ53″ | OP081818 | OP081825 |
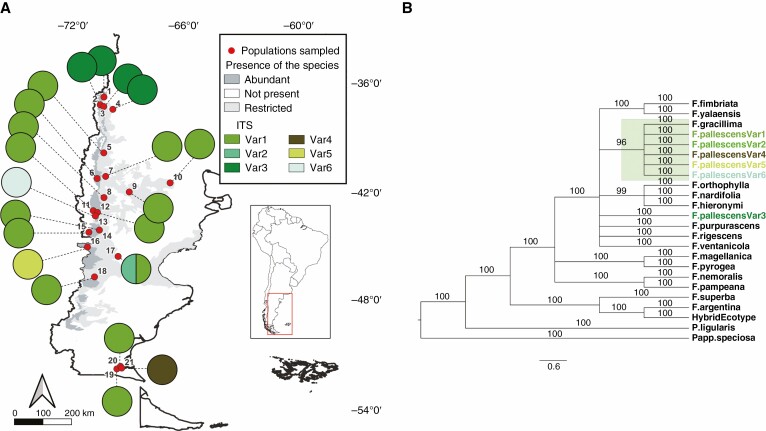
Fig. 1.
(A) Geographic distribution of the ITS variants of the evaluated populations of F. pallescens. Festuca pallescens variants are coloured in shades of green. (B) Bayesian tree based on nuclear ITS markers. Numbers above the branches indicate posterior probability values. The scale bar shows the expected substitutions per site. Pappostipa speciosa var. speciosa and Poa ligularis were used as outgroups.
DNA extraction and PCR amplification
DNA from at least three individuals per population was extracted. Leaf tissue was frozen using liquid nitrogen and ground to fine powder with an automatic mixer mill (Resch, Germany). DNA extraction followed the protocol of Doyle and Doyle (1987), with slight modifications (Gonzalo-Turpin and Hazard, 2009). Two DNA markers were used for the phylogenetic analysis: the internal transcribed spacer (ITS) and a chloroplast DNA region (trnL-F). The complete ITS region (ITS1-5.8S-ITS2) was amplified using the primers CY1-CY3 (Wright et al., 2006), ~600–700 bp in length (Torrecilla and Catalán, 2002; Catalán et al., 2004). For the amplification we used 40 ng of DNA as template, 0.625 U of GoTaq DNA polymerase (Promega, Madison, WI, USA) with 1× Colorless GoTaq® reaction buffer (Promega), 1.5 mm of MgCl2, 0.25 mm of each dNTP and 0.3 µm of each primer in a total volume of 30 µL. PCR reactions were carried out following this program: 4:30 min at 95 °C, 30 cycles of 30 s at 94 °C, 1 min at 56 °C and 2 min at 72 °C, and a final extension of 10 min at 72 °C. The trnL-F region (~1000 bp, Catalán et al., 2004) was amplified using the universal primers c and f (Taberlet et al., 1991) with 100 ng of DNA, 1 U of GoTaq DNA polymerase (Promega, Madison, WI, USA) with 1× Colorless GoTaq® reaction buffer (Promega), 2 mm MgCl2, 0.2 mm of each dNTP and 0.2 µm of each primer in a total volume of 50 µL. The amplification program used for this marker was as follows: 1 min at 94 °C followed by 35 cycles of 1 min at 94 °C, 1 min at 50 °C and 1 min at 72 °C, followed by a final extension of 7 min at 72 °C. PCR products for both markers were checked for positive amplification in 1.5 % agarose gels, stained with Gel Red, and visualized with a UV transilluminator. Amplified regions yielded bands of ~700 bp (ITS) and 1000 bp (trnL-F), which were purified using the ExoSAP-IT™ PCR Product Cleanup (Thermo Fisher) commercial kit and then sequenced in a capillary sequencer (ABI 3700, Unidad de Genómica, Instituto de Biotecnología de INTA, Hurlingham, Argentina).
Phylogenetic analyses
The ITS sequences were aligned using Muscle with manual adjustments when needed in AliView 1.26 (Larsson, 2014). For the phylogenetic analysis of the ITS region we additionally used sequences from 14 Patagonian species of Festuca retrieved from GenBank (Supplementary Data Table S1). In addition, a sequence from the ecotype of F. argentina previously mentioned was also included (López et al., 2018). Two Patagonian grasses (Poa ligularis var. ligularis and Pappostipa speciosa var. speciosa) were used as outgroups. All the ITS and trnL-F sequences of F. pallescens and F. gracillima were deposited in GenBank (accession numbers are provided in Supplementary Data Table S1).
A matrix of 24 sequences of 578 characters each was generated for the analysis of the ITS region. Gaps were treated as missing data and indels as point mutations. Boundaries of each sequence were established by alignment with the species of Festuca retrieved from GenBank (14 species). To assess the phylogenetic relationships between species of Patagonia and among populations of F. pallescens we performed analyses based on Bayesian inference (BI), maximum parsimony (MP) and maximum likelihood (ML) criteria. The IB, MP and ML analyses were carried out with MrBayes 3.2.7a (Ronquist et al., 2012), TNT 1.5 (Goloboff and Catalano, 2016) and the online software RAxML (Kozlov et al., 2019), respectively. We used JModelTest 2.1.10 to establish the best-fitting nucleotide substitution model that is required to run BI and ML analyses (Darriba et al., 2012). This resulted in model GTR + G according to both the Akaike information criterion (AIC) and Bayesian information criterion (BIC). Bayesian analyses were carried out by running pre-established parameters (1 million generations initiated from different random trees, sampling every 100th generation model parameters such as nucleotide substitution rates, γ shape, proportion of invariable sites, nucleotide frequency) estimated by MrBayes. After burn-in (discarding 25 % of the total sampling), parameters were sampled when reaching stationarity. The ITS consensus tree, obtained from 15 000 trees, was supported by branch values from the posterior probability. Clades with >95 % posterior probability values are considered well supported. Parsimony analyses were executed by subjecting the data set to a heuristic search strategy to find all equally parsimonious trees (MULPARSon, TBR) with 10 000 replicates, saving no more than ten trees of length equal to or shorter than 10 (López et al., 2018). These saved trees were collapsed to a consensus tree that was used as a negative constraint for a second search. All parsimonious trees obtained were used to compute a strict and 50 % majority rule consensus tree. Bootstrap support for branches was calculated through heuristic searches of 1000 replicates. Clades with bootstrap values <50 are shown unresolved. We used the software FigTree (http://tree.bio.ed.ac.uk/software/figtree/) to edit and visualize the consensus tree. To reconstruct the relationships between haplotypes we ran the program Network 10.2.0.0, which works with a median-joining network algorithm (Bandelt et al., 1999). The dataset used for the reconstruction contained the chloroplast DNA sequences of F. pallescens (seven haplotypes), F. gracillima, F. purpurascens and F. argentina and the sequence corresponding to the hybrid ecotype (López et al., 2018).
Morphological data
Twelve populations of F. pallescens that differ in either of the two evaluated molecular markers were selected for morphological characterization (Table 1). Plant material was obtained from the living collection of F. pallescens populations held at the Campo Experimental Agroforestal Trevelin (CEAT INTA Esquel; 43°07ʹ30″S, 71°33ʹ04″W, 356 masl), Argentina. Twenty-four quantitative and nine qualitative morphological traits were measured in 6–12 specimens of each population (Supplementary Data Table S2). Morphological characters included in this study are those traditionally used for the delimitation of species in Festuca (Dubcovsky and Martínez, 1988; Catalán and Müller, 2012). In this genus, the distribution of sclerenchyma in the leaf cross-section is a main trait for species identification (Aiken et al., 1985; Dubcovsky and Martínez, 1988). Therefore, leaf anatomy characters were observed in the middle region of the blade of the penultimate leaf of the short internodes zone in all specimens studied. The identification of different combinations of adaxial and/or abaxial girders of sclerenchyma of vascular bundles was based on leaf cross-sections stained with safranin.
Statistical analysis
The quantitative variables were tested for normality with the Shapiro–Wilks test (Mahibbur and Govindarajulu, 1997), as well as using diagram boxes. Homogeneity of variance was checked with Bartlett’s test. One-way ANOVA (α = 0.05) was performed to evaluate the significance of the differences among the specimens for each trait. Tukey’s test (P < 0.05) was applied for a posteriori comparison of each pair of means. For the analysis of the variable lemma awn length, population ACA (1) was excluded because plants lack awns in the lemma (see Results section).
The morphological data set (Supplementary Data Table S3) was analysed by discriminant and cluster analyses, with each character represented by mean values. Morphological character values were standardized prior to use in multivariate analyses. Pearson and Spearman correlation coefficients were estimated to identify pairs of highly correlated characters that may distort multivariate analyses (Michener and Sokal, 1957; Conover, 1999). Discriminant analysis allows samples to be classified within predefined groups using the discriminant functions adjusted for maximizing the between-groups to within-groups ratio of variance. Cluster analysis was performed to reveal the structure residing in the morphological dataset. Sample similarities using the morphological data matrix were calculated based on the Euclidian distance, and the average linkage hierarchical agglomerative method was used to establish clusters. The statistical analyses were performed using Infostat v. 2015 (Di Rienzo et al., 2015).
RESULTS
Variation at the ITS region
Six variants were identified at the ITS region along the distribution range of F. pallescens. The most common variant (F.pallescensVar1) was detected in 14 populations. Four northern populations [ACA (1), LMI (2), CHO (3) and HUI (4)] shared a distinctive ITS sequence, and the other four variants were found in two western populations [CRO (13) and FON (16)], one eastern population [RMA (17)], and one southern population [GAI (21)] (Fig. 1A). The ITS dataset included 19 taxa: F. pallescens (with six variants), 16 native species of Festuca, including the hybrid ecotype of F. argentina (Supplementary Data Table S1), and two outgroups (P. ligularis and P. speciosa). From the 578 aligned nucleotide positions, 73 (12.63 %) were parsimony-informative. The three phylogenetic analyses exhibited similar topologies, with two major clades that separate the F. pallescens variants from the hybrid ecotype. The BI tree showed the highest support values (Fig. 1A); MP and ML can be found in Supplementary Data Fig. S1. The ITS variants found in F. pallescens specimens formed a well-supported clade, associated with F. gracillima. A strong association with other Patagonian native species (e.g. F. purpurascens) and with northern species (e.g. F. ventanicola) was also observed. In a separate clade, the hybrid ecotype variant clustered together with F. argentina. This clade is sister to only one species distributed in the extreme north of Argentina (F. superba).
Variation in the chloroplast DNA (trnL-F) region
A total of seven chloroplast haplotypes were found among the analysed populations. The topology of the network shows the ancestral character of the most frequent haplotype (Hp1) (Fig. 2). Haplotype 4 shares the same six-nucleotide insertion as F. purpurascens but differs by two nucleotides in other positions. Haplotypes 2 and 5 share an insertion of 22 nucleotides but differ at two positions. One of these polymorphisms is an ambiguity (W in IUPAC code), with Hp2 showing W, and two alternative nucleotides observed in Hp1, Hp5, Hp6 and Hp7 (T), and Hp3 and Hp4 (A). Haplotype 7 shows high similarity with F. gracillima’s haplotype and is distanced from Hp1 by nine mutations. Additionally, the sympatric species F. argentina differs from Hp1 by 13 mutated positions and shares the same haplotype as the hybrid ecotype (Fig. 2).
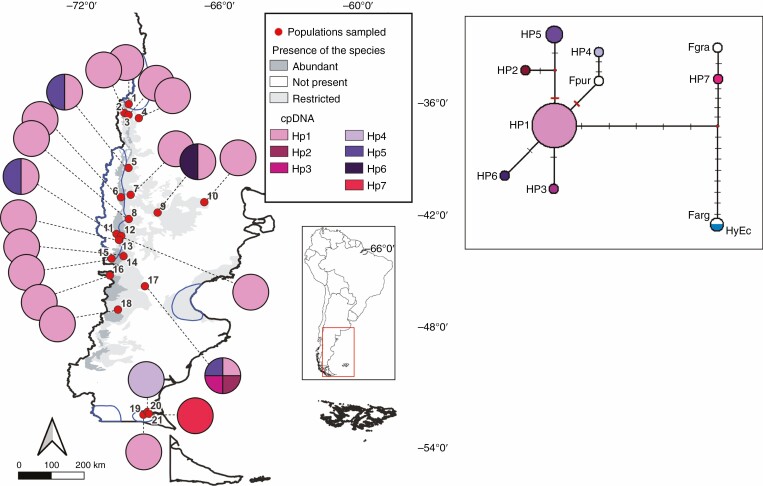
Fig. 2.
Geographic distribution of the haplotypes detected in F. pallescens populations along the entire distribution range. Putative glacial refugia [lowland, peripheral and valley refugia as described by Sérsic et al. (2011)] for different plant species are delimited with blue lines. The box on the right shows the haplotype network. Haplotypes are represented with different-sized circles according to their frequencies (HyEc, hybrid ecotype; Fgra, F. gracillima; Fpur, F. purpurascens; Farg, F. argentina). Point mutations are shown as black lines and indels as red lines.
The most frequent haplotype (Hp1) was found in 18 populations (Fig. 2). Haplotypes were fixed in 17 populations, 15 exhibiting the common haplotype and the other two [BLE (20) and GAI (21)] exclusive types. Intrapopulation variation was observed in four populations [CLI (5), JAC (9), YAG (11) and RMA (17)] (Fig. 2).
Morphological analyses
Significant differences between two or more populations for 20 of the 24 continuous variables were detected (ANOVA; P < 0.05). The average values and standard deviations of the quantitative traits analysed, as well as the results of a posteriori tests, are summarized in Supplementary Data Table S2. Population ACA (1), from northern Patagonia, was clearly the most differentiated. Its specimens showed ciliated ligules with significantly shorter hairs (P < 0.05). Northern populations [ACA (1), CLI (5), PIL (7), HUI (4) and JAC (9)] presented shorter hairs in the abaxial surface of leaves compared with southern populations [APE (12), CRO (13), FON (16), RMA (17), LBA (18), BLE (20) and GAI (21)]. Specimens of populations ACA (1) and HUI (4) presented longer ligules and were different compared with other populations (P < 0.05). In addition, populations ACA (1), HUI (4), CLI (5) and JAC (9) showed significantly shorter spikelets (P < 0.0001). Two populations of south Patagonia [LBA (20) and GAI (21)] had longer synflorescences (P < 0.0001). Plants from the CRO (13) and FON (16) populations presented longer lodicules (P < 0.0001).
Most variables were not correlated, except for spikelet length with lemma length (Pearson = 0.83), spikelet length with paleae length (Pearson = 0.87), and lemma length with paleae length (Pearson = 0.9). Pearson and Spearman correlation values were similar. Since the use of highly correlated characters is an implicit weighting of these characters and suggests potential multicollinearity problems, the total number of variables was reduced to 30 for final analyses (Supplementary Data Table S3).
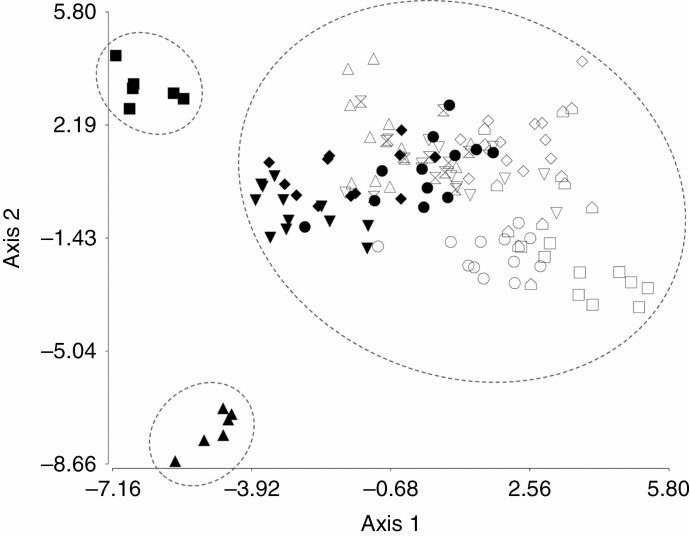
Discriminant analysis led to the identification of morphological patterns that allowed a good discrimination among some populations. Populations ACA (1) and HUI (4) were clearly discriminated when the first two discriminant functions (axis 1 and axis 2) were plotted (Fig. 3), while ACA (1), HUI (4) and CLI (5) were discriminated considering the axes 1 and 3 (Supplementary Data Fig. S2). The first three axes accounted for 62.82 % of the variation. The absolute values of the coefficients of the standardized discriminant functions are shown in Supplementary Data Table S3.
Fig. 3.
Scatterplot of scores derived from discriminant functions axis 1 and axis 2 produced by discriminant analysis applied to 30 morphological variables for the 12 populations of Festuca pallescens studied: ACA (solid squares), CLI (solid inverted triangles), PIL (solid diamonds), HUI (solid triangles), JAC (solid circles), CRO (open triangles), APE (open inverted triangles), GAI (open diamonds), LBA (open squares), RMA (open circles), FON (open pentagon), and BLE (open hourglasses).
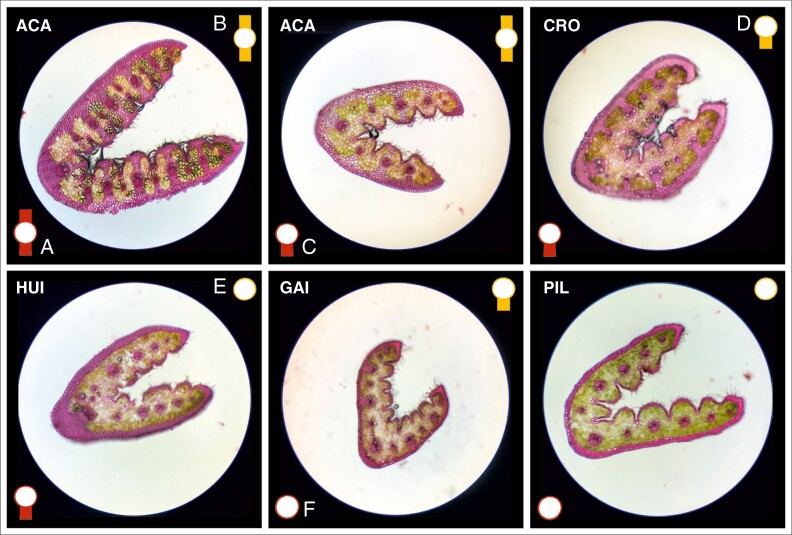
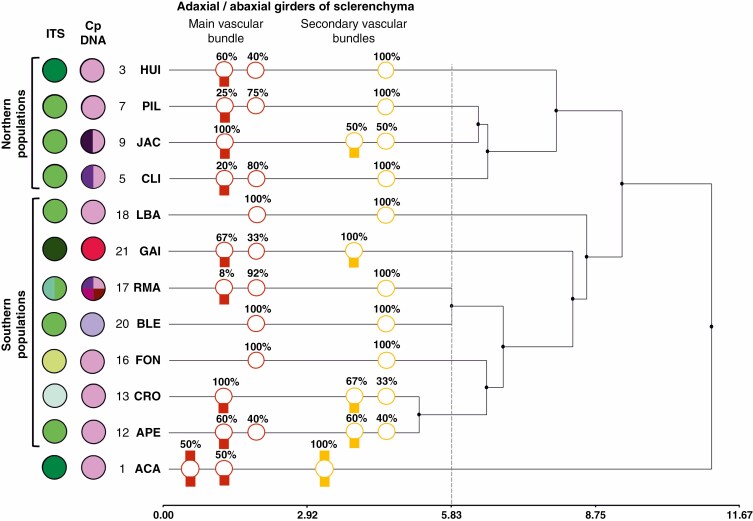
Leaf cross-sections revealed the presence of sclerenchyma under the abaxial epidermis and in the ribs under the adaxial epidermis in all specimens analysed. In addition, different combinations of adaxial and/or abaxial girders of sclerenchyma in vascular bundles were observed, depending on the population (Fig. 4). Population ACA (1) was the only one with double girders of sclerenchyma (adaxial to abaxial) in the main and secondary vascular bundles. In other populations the main and secondary vascular bundles showed different combinations with presence and/or absence of abaxial girders of sclerenchyma. In population ACA (1) all specimens showed abaxial girders of sclerenchyma in the main vascular bundle and 50 % of them presented double girders. In secondary vascular bundles all plants also presented double girders of sclerenchyma. Populations that presented abaxial girders of sclerenchyma in the main vascular bundle were HUI (4), CLI (5), PIL (7), JAC (9), APE (12), CRO (13), RMA (17) and GAI (21) (see different percentages in Fig. 5). In populations FON (16), LBA (18) and BLE (20), girders of sclerenchyma in the main vascular bundle were not observed. Populations that presented abaxial girders of sclerenchyma in secondary vascular bundles were PIL (7), JAC (9), APE (12), CRO (13) and GAI (21) (see percentages in Fig. 5). In populations HUI (4), CLI (5), FON (16), RMA (17), LBA (18) and BLE (20), girders of sclerenchyma in secondary vascular bundles were not present.
Fig. 4.
Leaf cross-section of specimens stained with safranin showing examples of the different combinations of adaxial and/or abaxial girders of sclerenchyma of vascular bundles. Population names follow those of Supplementary Data Table S1. (A) Adaxial to abaxial girders of sclerenchyma in main vascular bundle. (B) Adaxial to abaxial girders of sclerenchyma in secondary vascular bundles. (C) Abaxial girders of sclerenchyma in main vascular bundle. (D) Abaxial girders of sclerenchyma in secondary vascular bundles. (E) Secondary vascular bundles without girders of sclerenchyma. (F) Main vascular bundle without girders of sclerenchyma.
Fig. 5.
Dendrogram resulting from cluster analysis obtained with the morphological data of F. pallescens populations. The dendrogram includes morpho-anatomical results of leaf cross-sections (following Fig. 4) and molecular variation according to ITS and cpDNA. The scale corresponds to Euclidean distance.
In the cluster analysis, populations were grouped in terms of their similarity according to the 30 morphological traits analysed (Supplementary Data Table S3). Considering a reference line equal to 50 % of the maximum distance (a frequently used criterion) to validate clusters, the cut-off line was established at distance 5.83. The dendrogram shows that population ACA (1) is clearly separated from the rest and populations of north and south Patagonia were grouped into two main clusters (Fig. 5).
DISCUSSION
Maintaining genetic diversity across populations of a species is essential to ensure its survival, particularly under the current climate change context. We took the first steps towards describing differentiation among populations of a widespread keystone species of an arid and semiarid region. In this study, we found intraspecific variation at molecular markers and morpho-anatomical traits. Edge populations of F. pallescens hold different genetic variants in comparison with those located at the centre of the distribution. Distinct populations can be observed particularly at northern, eastern and southern sites. In general, the same pattern is observed for both markers. Similarly, morphological traits also separated populations into two latitudinal groups.
Intraspecific phylogeny
The Poaceae family includes >700 genera of economic and ecological significance, but phylogenetic relationships among species are complex and sometimes unresolved (Grass Phylogeny Working Group, 2001; Soreng et al., 2017). In addition, phylogenetic studies encompassing the relationships among Patagonian fescues are limited (Dubcovsky and Martínez, 1988; Ospina González, 2016; López et al., 2018). The genus Festuca holds >500 species all over the world (Inda et al., 2008). Although well supported phylogenies exist, the representation of Patagonian species is very scarce, e.g. only F. argentina, F. subantarctica, F. purpurascens, F. gracillima, F. magellanica and F. pyrogea were included (Torrecilla and Catalán, 2002; Torrecilla et al., 2003; Catalán et al., 2004; Inda et al., 2008; Minaya et al., 2017). In a previous study, we reported the position of F. pallescens within these phylogenies, clearly positioned within the ‘fine-leaved lineage’ (López et al., 2018). Here, with an enlarged sampling, we reconstructed the relationships between the Patagonian fescue species and the F. pallescens variants. Our results show that most of F. pallescens’ variants are closely related to F. gracillima, a sympatric species that occurs at the southernmost end of the distribution. This species is described as a complex together with F. pallescens (Dubcovsky and Martínez, 1991; Torrecilla and Catalán, 2002; Catalán and Müller, 2012). The variant detected in the four northernmost populations is separated from the main clade, forming an unresolved polytomy to another sympatric species, F. purpurascens. The natural distribution of F. purpurascens overlaps with F. pallescens’ distribution, and the two species showed high genetic similarities at both molecular markers.
Festuca gracillima, F. purpurascens and all the variants of F. pallescens were grouped with most species of Festuca, and were separated, with strong support, from a group that included F. argentina, F. superba and the hybrid ecotype. These results are consistent with Catalán et al. (2004), in which the festucoid grasses are separated into two major linages: ‘broad-leaved’ and ‘fine-leaved’ (Inda et al., 2014). Festuca argentina would belong to the first group, while F. purpurascens and F. pallescens would presumably belong to the latter (Catalán et al., 2004; Inda et al., 2008; López et al., 2018). In a further study of molecular, anatomical and micro-morphological characters, these species are also separated in different groups (Ospina González, 2016), but F. argentina is placed in an ambiguous position, close to either broad- or fine-leaved lineages. Even though Ospina González (2016) placed F. purpurascens and F. pallescens in a fine-leaved group according to his molecular results, anatomically he describes F. purpurascens within the broad-leaved lineage while F. pallescens is placed in the fine-leaved lineage together with F. gracillima. Clearly the high variation within and among species of the Festuca genus deserves more attention and further studies will contribute to a better comprehension of the relationships of the species belonging to this genus.
Genetic differentiation along the natural range
We identified eight distinct populations that differed from the most frequent ITS variant. Four of these populations are located at the northern edge of the distribution, and all the individuals sampled share the same sequence. This region was affected by sea introgressions and glaciations during the Miocene and Pleistocene (Ramos, 1982; Folguera and Ramos, 2002; Rabassa et al., 2005), which makes it a perfect spot for stochastic factors to develop differentiation from the central populations (Eckert et al., 2008). Within this northern region the extent of glaciations was limited to the foothills of the Andes (Flint and Fidalgo, 1964; Hollin and Schilling, 1981), leaving many ice-free areas where species could survive in situ (e.g. Markgraf et al., 1995; Sérsic et al., 2011; Soliani et al., 2012). At this latitude, F. pallescens shares its habitat with other native species of the genus, which are morphologically similar (e.g. F. acanthophylla var. acanthophylla and F. acanthophylla var. scabriuscula). Marginal populations are considered active regions for speciation processes which are particularly common in plants (Crawford, 2010; Rajakaruna, 2018). Ancient hybridization and speciation processes at glacial refugia might be a possible explanation of the variants shared by northern populations, but also of the southern variants found in this study (see next section).
Four populations at the species range edges have shown distinct variants. The occurrence of differentiation at suboptimal locations could imply an evolutionary advantage under extreme climatic conditions, allowing the species to colonize and remain at these further frontiers. For instance, plant species increase their ploidy level with latitude (Rice et al., 2019) and most Patagonian fescues are mainly polyploids (Dubcovsky and Martínez, 1988, 1992). Polyploidy diversifies an organism’s genetic background and provides selective advantages in novel or stressful environments (Van de Peer et al., 2017), and can also be a common way of sympatric speciation through hybridization processes (Wei et al., 2019; Gordon et al., 2020). Therefore, showing distinct genetic and morpho-anatomical variants, edge populations of F. pallescens could either suggest an ecological advantage (i.e. drought or salinity stress) or reinforce the possibility of hybridization with sympatric species.
A similar pattern of distribution of the genetic variation was displayed at the chloroplast level. The less frequent haplotypes found in our study were detected in marginal populations (east and south of the distribution). The populations carrying the alternative variants match the distribution of different putative glacial refugia described for Patagonian plant species (Cosacov et al., 2010; Sérsic et al., 2011; Baranzelli et al., 2022) (Fig. 2). Two haplotypes found in the southernmost populations near one of the aforementioned refugia (Sérsic et al., 2011) are of particular interest: Hp4 and Hp7 from populations BLE (20) and GAI (21), respectively. The haplotype 4 shares a step mutation with one haplotype found in population RMA (17), closer to the centre of the natural range for the species, but it also presents a six-nucleotide insertion that matches the sequence of F. purpurascens. On the other hand, Hp7 is almost an exact match of F. gracillima’s sequence. The populations BLE (20) and GAI (21) belong to the southernmost region of the distribution, where it overlaps with the distribution range of F. purpurascens and F. gracillima (Dubcovsky and Martínez, 1988; Ospina González et al., 2015). Hybridization with other species of the genus that share the same ploidy count is plausible and it has been argued as either a cause or consequence of speciation (Dubcovsky and Martínez, 1991, 1992; Oliva, 1996; Šmarda and Stančík, 2005; Ospina González, 2016; Fernández et al., 2017). Fescues are known to hybridize within the genus (Humphreys et al., 1995; Inda et al., 2014), and a putative hybrid ecotype has already been described in a location with suboptimal conditions and isolated from the centre of the distribution (López et al., 2018). Interspecific hybrids have also been described as common among other species of the Poaceae, and these events can complicate the reconstruction of phylogenetic relations between species (Díaz-Pérez et al., 2014; Baiakhmetov et al., 2020).
The high genetic diversity detected in population RMA (17) at both markers deserves attention. This population is close to a described plant refuge and a phylogeographical break for several species reviewed by Sérsic et al. (2011). The steppe vegetation is poorly represented since most studies were devoted to temperate forest species. However, in situ survival throughout glacial cycles was suggested based on phylogeography and ecological niche modelling that indicated stable geographical distribution of two Hordeum species for at least the Holocene (Jakob et al., 2009). Past vegetation reconstruction within the southern distribution range of F. pallescens confirms its presence during the Holocene (Echeverría et al., 2022). The legacy of the glacial era is still imprinted in the current taxa, but F. pallescens was never studied. Our results are the first information contributing to knowledge of the evolutionary history of the species. Future studies should include other chloroplast markers to infer phylogeographical patterns.
Morphological and anatomical comparisons
In the genus Festuca there are vegetative characters that are of great taxonomic importance and one of them is the internal morphology of the leaves (Saint-Yves, 1927; Parodi, 1953; Aiken et al., 1985). In Patagonian fescues, the distribution of sclerenchyma on the leaves not only helps to explain the geographical distribution of some of the species but also taxonomically separates the species into different groups (Dubcovsky and Martínez, 1988). Festuca pallescens can be distinguished by the presence of sclerenchyma under the abaxial epidermis and in the ribs under the adaxial epidermis, but does not possess girders of sclerenchyma in the main and secondary vascular bundles (Dubcovsky and Martínez, 1988). The morphological analyses allowed us to confirm the taxonomic identity of the studied specimens and, in addition, to detect the phenotypic variability of the species throughout its natural distribution range.
The specimens from ACA population (1) were morpho-anatomically the most different among all populations analysed. Unlike the other populations, plants in ACA presented awnless lemmas, double girders of sclerenchyma (adaxial to abaxial) in the main and secondary vascular bundles and ligules with the shortest hairs. According to Dubcovsky and Martínez (1988), these characteristics are observed in F. purpurascens, F. argentina, F. acanthophylla var. acanthophylla, F. cirrosa, F. monticola and F. acanthophylla var. scabriuscula. Within the region of the ACA population, F. pallescens grows in sympatry with F. acanthophylla var. scabriuscula and var. acanthophylla. The specimens of the ACA population presented other leaf anatomical traits (e.g. number of vascular bundles and the arrangement of subepidermal sclerenchyma) that are more similar to those typical of these two species than to F. pallescens. Similarly, the southernmost populations [BLE (20) and GAI (21)] live in sympatry with F. gracillima. Although leaf anatomy in BLE specimens is similar to that in F. gracillima, that in GAI is not, because they present abaxial girders in the main and secondary vascular bundles. As previously stated for genetic variability, these results might be evidence of ancient hybridization processes that occurred during the Pleistocene at periglacial refugia at both the northern and southern limits of the distribution range. Different putative glacial refugia have been described within the current distribution range of F. pallescens, from north-west Patagonia (~35° S) to the south of Santa Cruz (~51° S) (Sérsic et al., 2011). Moreover, the distribution ranges of F. acanthophylla and F. purpurascens in the north and F. gracillima in the south match with some of these areas (Dubcovsky and Martínez, 1988) (Fig. 2). The coexistence of these species with F. pallescens in confined proximity and through many recolonization cycles could have promoted interspecific crossing. Hybridization during prolonged isolation in glacial times was reported for other species around the world (e.g. Consaul et al., 2010; Klein and Kadereit, 2016; Liu et al., 2018; Shepherd et al., 2022) and also in Patagonia (e.g. Tremetsberger et al., 2009; Soliani et al., 2012; Azpilicueta et al., 2014).
A latitudinal pattern in the distribution of genetic variation is frequently found within Patagonian species, both plants and animals, mainly related to historical processes like glaciations (e.g. Bekessy et al., 2002; Marchelli and Gallo, 2006; Azpilicueta et al., 2009, 2014; Arana et al., 2010; Cosacov et al., 2010; Lessa et al., 2010; Mathiasen and Premoli, 2010; Sérsic et al., 2011; Soliani et al., 2012, 2015; Vera-Escalona et al., 2012; Baranzelli, 2017; Fasanella et al., 2017; Renny, 2022). The variation detected in our study indicates that F. pallescens would follow the same latitudinal pattern. Northern populations shared some traits that differed from those recorded in the southern populations. For example, the ACA (1) and HUI (4) populations presented the longest ligules. In addition, the populations of northern Patagonia [ACA, HUI, CLI (5), PIL (7) and JAC (9)] differed from the other populations by presenting significantly shorter hairs on the abaxial surface of leaves. Moreover, the ACA, HUI, CLI and JAC populations presented significantly shorter spikelets. Discriminant analyses separated the ACA, HUI and CLI populations, leaving the rest of the populations without a defined grouping pattern. From the results of the cluster analysis, it is clear that the ACA population is separate from the rest of the populations, which grouped into two main subgroups according to their latitudinal geographical distribution (north and south populations).
Conclusions
Our results showed a low genetic differentiation among central populations but distinctiveness at peripheral sites, suggesting that hybridization of F. pallescens with sympatric species is possible; morphological traits and molecular markers converged on similar outcomes. Low genetic variability is an expected outcome of permanent persistence in situ as suggested by Jakob et al. (2009) for Hordeum spp. Pollen and phytolith records showed evidence of the presence of current steppe species belonging to the Poaceae family during the early Quaternary, reinforcing this possibility (Palazzesi and Barreda, 2012; Palazzesi et al., 2021). In the north, the group of analysed populations shared the same distinct molecular variants but displayed differences in morphological traits. Hybridization with sympatric species at these latitudes may be suggested, and even different stages of speciation can be proposed given the high separation of the ACA population from the rest of the northern populations. This population is at the limit of the distribution range. A similar situation can be inferred at the southern edge, where populations resemble those of F. gracillima in both molecular and morphological characters. Marginal sites usually offer suboptimal conditions for a species; therefore, hybridization can play a key role in adaptation (Španiel and Rešetnik, 2022).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to A. Beider, A. Mogni, E. Vivar, G. Oliva, G. Siffredi, H. Morgana, M. Easdale and W. Opazo for helping with the collection of biological material used for this study. We specially thank F. Umaña for the assistance in the use of QGIS for the preparation of Figs 1 and 2, and R. Vidal Russell for the assistance in the use of TNT software. Sampling permissions were provided by the corresponding provinces (Neuquén, Río Negro, Chubut and Santa Cruz, Argentina).
Contributor Information
V Guidalevich, INTA Bariloche – IFAB (INTA-CONICET), Modesta Victoria 4450, 8400, Bariloche, Argentina.
N Nagahama, EEAf Esquel INTA, Chacabuco 513, 9200, Esquel, Argentina.
A S López, INTA Bariloche – IFAB (INTA-CONICET), Modesta Victoria 4450, 8400, Bariloche, Argentina.
J P Angeli, EEAf Esquel INTA, Chacabuco 513, 9200, Esquel, Argentina.
P Marchelli, INTA Bariloche – IFAB (INTA-CONICET), Modesta Victoria 4450, 8400, Bariloche, Argentina.
M M Azpilicueta, INTA Bariloche – IFAB (INTA-CONICET), Modesta Victoria 4450, 8400, Bariloche, Argentina.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: species and GenBank accession numbers. Table S2: one-way ANOVA. Table S3: discriminant analysis. Figure S1: maximum parsimony and likelihood tree based on nuclear data set. Figure S2: scatterplot of scores derived from axis 1 versus axis 3 produced by discriminant analysis applied to 30 morphological variables for the 12 populations studied.
FUNDING
This study was financed by Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación through PICT 2017-2379, Instituto Nacional de Tecnología Agropecuaria through PE I142 and Consejo Nacional de Investigaciones Científicas y Técnicas through PUE CONICET 069. V.G. was supported by a fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas and J.P.A. by a cofinanced fellowship from Instituto Nacional de Tecnología Agropecuaria and Consejo Nacional de Investigaciones Científicas y Técnicas.
LITERATURE CITED
- Aiken SG, Darbyshire SJ, Lefkovitch LP.. 1985. Restricted taxonomic value of leaf sections in Canadian narrow-leaved Festuca (Poaceae). Canadian Journal of Botany 63: 995–1007. doi: 10.1139/b85-135. [DOI] [Google Scholar]
- Arana MV, Gallo LA, Vendramin GG, Pastorino MJ, Sebastiani F, Marchelli P.. 2010. High genetic variation in marginal fragmented populations at extreme climatic conditions of the Patagonian cypress Austrocedrus chilensis. Molecular Phylogenetics and Evolution 54: 941–949. ISSN 1055-7903. doi: 10.1016/j.ympev.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Azpilicueta MM, Marchelli P, Gallo LA.. 2009. The effects of Quaternary glaciations in Patagonia as evidenced by chloroplast DNA phylogeography of southern beech Nothofagus obliqua. Tree Genetics and Genomes 5: 561–571. doi: 10.1007/s11295-009-0209-x. [DOI] [Google Scholar]
- Azpilicueta MM, Pastorino M, Puntieri J, et al. 2014. Robles in Lagunas de Epulauquen, Argentina: previous and recent evidence of their distinctive character. Revista Chilena de Historia Natural 87: 24–35. doi: 10.1186/s40693-014-0024-0. [DOI] [Google Scholar]
- Baiakhmetov E, Nowak A, Gudkova PD, Nobis M.. 2020. Morphological and genome-wide evidence for natural hybridisation within the genus Stipa (Poaceae). Scientific Reports 10: 13803. doi: 10.1038/s41598-020-70582-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A.. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48. doi: 10.1093/oxfordjournals.molbev.a026036. https://www.fluxus-engineering.com/sharenet.htm (28 September 2022). [DOI] [PubMed] [Google Scholar]
- Baranzelli MC, Cosacov A, Ferreiro G, Johnson LA, Sérsic AN.. 2017. Travelling to the south: phylogeographic spatial diffusion model in Monttea aphylla (Plantaginaceae), an endemic plant of the Monte Desert. PLoS One 12: e0178827. doi: 10.1371/journal.pone.0178827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranzelli MC, Cosacov A, Sede SM, Nicola MV, Sérsic AN.. 2022. Anthropocene refugia in Patagonia: a macrogenetic approach to safeguarding the biodiversity of flowering plants. Biological Conservation 268: 109492. doi: 10.1016/j.biocon.2022.109492. [DOI] [Google Scholar]
- Bekessy SA, Allnutt TR, Premoli AC, et al. 2002Genetic variation in the Monkey Puzzle tree, detected using RAPDs. Heredity 88: 243–249. doi: 10.1038/sj.hdy.6800033. [DOI] [PubMed] [Google Scholar]
- Bertiller MB, Bisigato A.. 1998. Vegetation dynamics under grazing disturbance. The state-and- transition model for the Patagonian steppes. Ecología Austral 8: 191–199. [Google Scholar]
- Bertiller MB, Irisarri MP, Ares JO.. 1990. Phenology of Festuca pallescens in relation to topography in north-western Patagonia. Journal of Vegetation Science 1: 579–584. doi: 10.2307/3235562. [DOI] [Google Scholar]
- Bradford JB, Schlaepfer DR, Lauenroth WK, Palmquist KA.. 2020. Robust ecological drought projections for drylands in the 21st century. Global Change Biology 26: 3906–3919. doi: 10.1111/gcb.15075. [DOI] [PubMed] [Google Scholar]
- Caballé G, Fernández ME, Gyenge J, Aparicio A, Schlichter T.. 2011. Modeling leaf maximum net photosynthetic rate of Festuca pallescens, the dominant perennial grass of Patagonian pine-based silvopastoral systems. Agroforestry Systems 83: 13–24. doi: 10.1007/s10457-011-9382-7. [DOI] [Google Scholar]
- Catalán P, Müller J.. 2012. Festuca L. In: Anton MA, Zuloaga FO, eds. Flora de Argentina, Vol. 3. Buenos Aires: CONICET, 219–250. [Google Scholar]
- Catalán P, Torrecilla P, Rodríguez JAL, Olmstead RG.. 2004. Phylogeny of the festucoid grasses of subtribe Loliinae and allies (Poeae, Pooideae) inferred from ITS and trnL–F sequences. Molecular Phylogenetics and Evolution 31: 517–541. doi: 10.1016/j.ympev.2003.08.025. [DOI] [PubMed] [Google Scholar]
- Conover WJ. 1999. Practical nonparametric statistics. New York: John Wiley and Sons. [Google Scholar]
- Consaul LL, Gillespie LJ, Waterway MJ.. 2010. Evolution and polyploid origins in North American Arctic Puccinellia (Poaceae) based on nuclear ribosomal spacer and chloroplast DNA sequences. American Journal of Botany 97: 324–336. doi: 10.3732/ajb.0900180. [DOI] [PubMed] [Google Scholar]
- Cosacov A, Sérsic AN, Sosa V, Johnson LA, Cocucci AA.. 2010. Multiple periglacial refugia in the Patagonian steppe and post-glacial colonization of the Andes: the phylogeography of Calceolaria polyrhiza. Journal of Biogeography 37: 1463–1477. doi: 10.1111/j.1365-2699.2010.02307.x. [DOI] [Google Scholar]
- Crawford DJ. 2010. Progenitor-derivative species pairs and plant speciation. Taxon 59: 1413–1423. doi: 10.1002/tax.595008. [DOI] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Poulsen L, Schulte-Herbrüggen B, et al. 2012. Conserving dryland biodiversity. Nairobi: International Union for the Conservation of Nature. [Google Scholar]
- Defossé GE, Bertiller MB, Ares JO.. 1990. Above-ground phytomass dynamics in a grassland steppe of Patagonia, Argentina. Journal of Range Management 43: 157. doi: 10.2307/3899036. [DOI] [Google Scholar]
- Díaz-Pérez AJ, Sharifi-Tehrani M, Inda LA, Catalán P.. 2014. Polyphyly, gene-duplication and extensive allopolyploidy framed the evolution of the ephemeral Vulpia grasses and other fine-leaved Loliinae (Poaceae). Molecular Phylogenetics and Evolution 79: 92–105. doi: 10.1016/j.ympev.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Doyle J, Doyle J.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Dubcovsky J, Martínez A.. 1988. Phenetic relationships in the Festuca spp. from Patagonia. Canadian Journal of Botany 66: 468–478. doi: 10.1139/b88-072. [DOI] [Google Scholar]
- Dubcovsky J, Martínez AJ.. 1991. Chromosome complement and nucleoli in the Festuca pallescens alliance from South America. Canadian Journal of Botany 69: 2756–2761. doi: 10.1139/b91-346. [DOI] [Google Scholar]
- Dubcovsky J, Martínez AJ.. 1992. Cytotaxonomy of the Festuca spp. from Patagonia. Canadian Journal of Botany 70: 1134–1140. doi: 10.1139/b92-140. [DOI] [Google Scholar]
- Echeverría MR, Bamonte FP, Marcos MA, Sottile GD, Mancini MV.. 2022. Past vegetation reconstruction maps and paleoclimatic variability inferred by pollen records in southern Patagonia Argentina since the Late Glacial-Holocene transition. Journal of South American Earth Sciences 116: 103834. doi: 10.1016/j.jsames.2022.103834. [DOI] [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC.. 2008. Genetic variation across species’ geographical ranges: the central–marginal hypothesis and beyond. Molecular Ecology 17: 1170–1188. doi: 10.1111/j.1365-294X.2007.03659.x. [DOI] [PubMed] [Google Scholar]
- Fasanella M, Premoli AC, Urdampilleta JD, González ML, Chiapella JO.. 2017. How did a grass reach Antarctica? The Patagonian connection of Deschampsia antarctica (Poaceae). Botanical Journal of the Linnean Society 185: 511–524. doi: 10.1093/botlinnean/box070. [DOI] [Google Scholar]
- Fernández ME, Gyenge JE, Schlichter TM.. 2004. Shade acclimation in the forage grass Festuca pallescens: biomass allocation and foliage orientation. Agroforestry Systems 60: 159–166. doi: 10.1023/b:agfo.0000013276.68254.78. [DOI] [Google Scholar]
- Fernández ME, Gyenge JE, Schlichter TM.. 2006. Growth of Festuca pallescens in silvopastoral systems in Patagonia, part 1: positive balance between competition and facilitation. Agroforestry Systems 66: 259–269. doi: 10.1007/s10457-005-0590-x. [DOI] [Google Scholar]
- Fernández M, Ezcurra C, Calviño CI.. 2017. Chloroplast and ITS phylogenies to understand the evolutionary history of southern South American Azorella, Laretia and Mulinum (Azorelloideae, Apiaceae). Molecular Phylogenetics and Evolution 108: 1–21. doi: 10.1016/j.ympev.2017.01.016. [DOI] [PubMed] [Google Scholar]
- Flint RF, Fidalgo F.. 1964. Glacial geology of the East flank of the Argentine Andes between latitude 39°10ʹS. and latitude 41°20ʹS. Geological Society of America Bulletin 75: 335–352. doi: 10.1130/0016-7606(1964)75[335:ggotef]2.0.co;2. [DOI] [Google Scholar]
- Folguera A, Ramos VA.. 2002. The effects produced by the approaching, collision and subduction of Pacific ridges in the Patagonian Andes. Acta Geologica Hispanica 37: 329–353. [Google Scholar]
- Gaitán JJ, Bran DE, Oliva GE, et al. 2017. Aridity and overgrazing have convergent effects on ecosystem structure and functioning in Patagonian rangelands. Land Degradation and Development 29: 210–218. doi: 10.1002/ldr.2694. [DOI] [Google Scholar]
- Gaitán JJ, Bran DE, Oliva GE.. 2020. Patagonian desert. In: Goldstein MI, DellaSalla DA, eds. Encyclopedia of the world’s biomes, Vol. 2. Elsevier, 163–180. doi: 10.1016/B978-0-12-409548-9.11929-3. [DOI] [Google Scholar]
- Goloboff P, Catalano S.. 2016. TNT, version 1.5, with a full implementation of phylogenetic morphometrics. Cladistics 32: 221–238. doi: 10.1111/cla.12160. [DOI] [PubMed] [Google Scholar]
- Golodets C, Sternberg M, Kigel J, et al. 2015. Climate change scenarios of herbaceous production along an aridity gradient: vulnerability increases with aridity. Oecologia 177: 971–979. doi: 10.1007/s00442-015-3234-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez SL, Ghermandi L.. 2021. Overgrazing causes a reduction in the vegetation cover and seed bank of Patagonian grasslands. Plant and Soil 464: 75–87. doi: 10.1007/s11104-021-04931-y. [DOI] [Google Scholar]
- Gonzalo-Turpin H, Hazard L.. 2009. Local adaptation occurs along altitudinal gradient despite the existence of gene flow in the alpine plant species Festuca eskia. Journal of Ecology 97: 742–751. doi: 10.1111/j.1365-2745.2009.01509.x. [DOI] [Google Scholar]
- Gordon SP, Contreras-Moreira B, Levy JJ, et al. 2020. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nature Communications 11: 3670. doi: 10.1038/s41467-020-17302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau HR, Aide M.. 2008. Globalization and land-use transitions in Latin America. Ecology and Society 13: 16. doi: 10.5751/ES-02559-130216. [DOI] [Google Scholar]
- Grass Phylogeny Working Group, Barker NP, Clark LG, et al. 2001. Phylogeny and subfamilial classification of the grasses (Poaceae). Annals of the Missouri Botanical Garden 88: 373. doi: 10.2307/3298585. [DOI] [Google Scholar]
- Hoffmann AA, Hercus MJ.. 2000. Environmental stress as an evolutionary force. BioScience 50: 217. doi: 10.1641/0006-3568(2000)050[0217:esaaef]2.3.co;2. [DOI] [Google Scholar]
- Hollin JT, Schilling DH.. 1981. Late Wisconsin-Weichselian mountain glaciers and small ice caps. In: Denton GH, Hughes TJ, eds. The last great ice sheets. New York: Wiley Interscience, 179–206. [Google Scholar]
- Hufford KM, Mazer SJ.. 2003. Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends in Ecology and Evolution 18: 147–155. doi: 10.1016/s0169-5347(03)00002-8. [DOI] [Google Scholar]
- Humphreys MW, Thomas HM, Morgan WG, et al. 1995. Discriminating the ancestral progenitors of hexaploid Festuca arundinacea using genomic in situ hybridization. Heredity 75: 171–174. doi: 10.1038/hdy.1995.120. [DOI] [Google Scholar]
- Inda LA, Segarra-Moragues JG, Müller J, Peterson PM, Catalán P.. 2008. Dated historical biogeography of the temperate Loliinae (Poaceae, Pooideae) grasses in the northern and southern hemispheres. Molecular Phylogenetics and Evolution 46: 932–957. doi: 10.1016/j.ympev.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Inda LA, Sanmartín I, Buerki S, Catalán P.. 2014. Mediterranean origin and Miocene-Holocene Old World diversification of meadow fescues and ryegrasses (Festuca subgenus Schedonorus and Lolium). Journal of Biogeography 41: 600–614. doi: 10.1111/jbi.12211. [DOI] [Google Scholar]
- Jakob SS, Martinez-Meyer E, Blattner FR.. 2009. Phylogeographic analyses and paleodistribution modeling indicate Pleistocene in situ survival of Hordeum species (Poaceae) in southern Patagonia without genetic or spatial restriction. Molecular Biology and Evolution 26: 907–923. doi: 10.1093/molbev/msp012. [DOI] [PubMed] [Google Scholar]
- James JJ, Sheley RL, Erickson T, Rollins KS, Taylor MH, Dixon KW.. 2013. A systems approach to restoring degraded drylands. Journal of Applied Ecology 50: 730–739. doi: 10.1111/1365-2664.12090. [DOI] [Google Scholar]
- Klein JT, Kadereit JW.. 2016. Allopatric hybrids as evidence for past range dynamics in Sempervivum (Crassulaceae), a western Eurasian high mountain oreophyte. Alp Botany 126: 119–133. doi: 10.1007/s00035-016-0164-8. [DOI] [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A.. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30: 3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessa EP, D’Elía G, Pardiñas UFJ.. 2010. Genetic footprints of late Quaternary climate change in the diversity of Patagonian-Fueguian rodents. Molecular Ecology 19: 3031–3037. doi: 10.1111/j.1365-294X.2010.04734.x. [DOI] [PubMed] [Google Scholar]
- Liu YP, Ren ZM, Harris AJ, Peterson PM, Wen J, Su X.. 2018. Phylogeography of Orinus (Poaceae), a dominant grass genus on the Qinghai-Tibet Plateau. Botanical Journal of the Linnean Society 186: 202–223. doi: 10.1093/botlinnean/box091. [DOI] [Google Scholar]
- Liu AT, Zou CJ, Zhang C, Zheng YR, Shimizu H, Xu WD.. 2011. Intraspecific diversity: adaptive differentiation of Picea mongolica WD Xu ecotypes. Forestry Studies in China 13: 189–197. doi: 10.1007/s11632-011-0307-3. [DOI] [Google Scholar]
- López AS, Azpilicueta MM, López DR, Siffredi GL, Marchelli P.. 2018. Phylogenetic relationships and intraspecific diversity of a north Patagonian fescue: evidence of differentiation and interspecific introgression at peripheral populations. Folia Geobotanica 53: 115–131. doi: 10.1007/s12224-017-9304-1. [DOI] [Google Scholar]
- López AS, Marchelli P, Batlla D, López DR, Arana MV.. 2019. Seed responses to temperature indicate different germination strategies among Festuca pallescens populations from semi-arid environments in North Patagonia. Agricultural and Forest Meteorology 27: 81–90. doi: 10.1016/j.agrformet.2019.04.002. [DOI] [Google Scholar]
- López AS, López DR, Caballé G, Siffredi GL, Marchelli P.. 2020. Local adaptation along a sharp rainfall gradient occurs in a native Patagonian grass, Festuca pallescens, regardless of extensive gene flow. Environmental and Experimental Botany 171: 103933. doi: 10.1016/j.envexpbot.2019.103933. [DOI] [Google Scholar]
- López AS, López DR, Arana MV, Batlla D, Marchelli P.. 2021. Germination response to water availability in populations of Festuca pallescens along a Patagonian rainfall gradient based on hydrotime model parameters. Scientific Reports 11: 10653. doi: 10.1038/s41598-021-89901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre FT, Benito BM, Berdugo M, et al. 2021. Biogeography of global drylands. New Phytologist 231: 540–558. doi: 10.1111/nph.17395. [DOI] [PubMed] [Google Scholar]
- Maestre FT, Salguero-Gómez R, Quero JL.. 2012. It is getting hotter in here: determining and projecting the impacts of global environmental change on drylands. Philosophical Transactions of the Royal Society B: Biological Sciences 367: 3062–3075. doi: 10.1098/rstb.2011.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrin GO, Marengo JA, Boulanger J-P, et al. 2014. Central and South America. In: Barros VR, Field CB, Dokken DJ, Mastrandrea MD, Mach KJ, Bilir TE, Chatterjee M, Levy AN, MacCracken S, Mastrandrea PR, White LL, eds. Climate Change 2014: Impacts, adaptation, and vulnerability. Part B: regional aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press, 1499–1566. [Google Scholar]
- Mahibbur RM, Govindarajulu Z.. 1997. A modification of the test of Shapiro and Wilks for normality. Joumal of Applied Statistics 24: 219–235. [Google Scholar]
- Manel S, Gugerli F, Thuiller W, et al.; IntraBioDiv Consortium. 2012. Broad-scale adaptive genetic variation in alpine plants is driven by temperature and precipitation. Molecular Ecology 21: 3729–3738. doi: 10.1111/j.1365-294X.2012.05656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchelli P, Gallo L.. 2006. Multiple ice-age refugia in a southern beech of South America as evidenced by chloroplast DNA markers. Conservation Genetics 7: 591–603. doi: 10.1007/s10592-005-9069-6. [DOI] [Google Scholar]
- Markgraf V, McGlone M, Hope G.. 1995. Neogene paleoenvironmental and paleoclimatic change in southern temperate ecosystems—a southern perspective. Trends in Ecology and Evolution 10: 143–147. doi: 10.1016/s0169-5347(00)89023-0. [DOI] [PubMed] [Google Scholar]
- Mathiasen P, Premoli AC.. 2010. Out in the cold: genetic variation of Nothofagus pumilio (Nothofagaceae) provides evidence for latitudinally distinct evolutionary histories in austral South America. Molecular Ecology 19: 371–385. doi: 10.1111/j.1365-294X.2009.04456.x. [DOI] [PubMed] [Google Scholar]
- Michener CD, Sokal RR.. 1957. A quantitative approach to a problem in classification. Evolution 11: 130–162. doi: 10.1111/j.1558-5646.1957.tb02884.x. [DOI] [Google Scholar]
- Minaya M, Hackel J, Namaganda M, et al. 2017. Contrasting dispersal histories of broad- and fine-leaved temperate Loliinae grasses: range expansion, founder events, and the roles of distance and barriers. Journal of Biogeography 44: 1980–1993. doi: 10.1111/jbi.13116. [DOI] [Google Scholar]
- Moreno L, Bertiller MB.. 2015. Phenotypic plasticity of morpho-chemical traits of perennial grasses from contrasting environments of arid Patagonia. Journal of Arid Environments 116: 96–102. doi: 10.1016/j.jaridenv.2015.02.007. [DOI] [Google Scholar]
- Nicora EG. 1978. Gramineae. In: Correa MN, ed. Flora Patagónica 3. Buenos Aires: Instituto Nacional de Tecnología Agropecuaria. 93–121. [Google Scholar]
- Oliva G. 1996. Biología de poblaciones de Festuca gracillima. PhD Thesis, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Argentina. [Google Scholar]
- Oliva G, dos Santos E, Sofía O, et al. 2020. The MARAS dataset, vegetation and soil characteristics of dryland rangelands across Patagonia. Scientific Data 7: 327. doi: 10.1038/s41597-020-00658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospina González JC. 2016. Estudios morfológicos, anatómicos, taxonómicos y relaciones filogenéticas de las especies de Festuca del Cono Sur (Argentina, Chile, Uruguay, Paraguay y sur de Brasil). PhD Thesis. Universidad Nacional de La Plata, Argentina. doi: 10.35537/10915/52004 [DOI] [Google Scholar]
- Ospina González JC, Aliscioni SS, Denham SS.. 2015. A revision of Festuca (Loliinae, Pooideae, Poaceae) in Chile. Phytotaxa 223: 1. doi: 10.11646/phytotaxa.223.1.1. [DOI] [Google Scholar]
- Palazzesi L, Barreda V.. 2012. Fossil pollen records reveal a late rise of open-habitat ecosystems in Patagonia. Nature Communications 3: 1294. doi: 10.1038/ncomms2299. [DOI] [PubMed] [Google Scholar]
- Palazzesi L, Vizcaíno SF, Barreda VD, et al. 2021. Reconstructing Cenozoic Patagonian biotas using multi-proxy fossil records. Journal of South American Earth Sciences 112: 103513. [Google Scholar]
- Parodi LR. 1953. Las especies de Festuca de la Patagonia. Revista Argentina de Agronomía 20: 177–229. [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K.. 2017. The evolutionary significance of polyploidy. Nature Reviews Genetics 18: 411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- Pourebrahimi S, Mirshamsi O, Ghasempouri SM, Moghaddam FY, Aliabadian M.. 2022. Phylogeny and evolutionary history of the sombre tit, Poecile lugubris in the western Palearctic (Aves, Paridae). Molecular Phylogenetics and Evolution 167: 107343. doi: 10.1016/j.ympev.2021.107343. [DOI] [PubMed] [Google Scholar]
- Rabassa J, Coronato AM, Salemme M.. 2005. Chronology of the Late Cenozoic Patagonian glaciations and their correlation with biostratigraphic units of the Pampean region (Argentina). Asian Journal of Earth Sciences 20: 81–103. [Google Scholar]
- Rajakaruna N. 2018. Lessons on evolution from the study of edaphic specialization. Botanical Review 84: 39–78. doi: 10.1007/s12229-017-9193-2. [DOI] [Google Scholar]
- Ramos VA. 1982. Las ingresiones del Terciario en el Norte de la Patagonia. In: III Congreso Geológico Chileno. Actas I(A), (Concepción), 262–288. [Google Scholar]
- Renny M, Acosta MC, Sérsic AN.. 2022. Ancient climate changes and Andes uplift, rather than Last Glacial Maximum, affected distribution and genetic diversity patterns of the southernmost mycoheterotrophic plant Arachnitis uniflora Phil. (Corsiaceae). Global and Planetary Change 208: 103701. doi: 10.1016/j.gloplacha.2021.103701. [DOI] [Google Scholar]
- Rice A, Šmarda P, Novosolov M, et al. 2019. The global biogeography of polyploid plants. Nature Ecology and Evolution 3: 265–273. doi: 10.1038/s41559-018-0787-9. [DOI] [PubMed] [Google Scholar]
- Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW.. 2015. InfoStat versión 2015. Córdoba:Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, et al. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Yves A. 1927. Contribution a l’etude des Festuca (subgen. Eu-Festuca) de l’Amerique du Sud. Candollea 3: 151–315. [Google Scholar]
- Sérsic AN, Cosacov A, Cocucci AA, et al. 2011. Emerging phylogeographical patterns of plants and terrestrial vertebrates from Patagonia. Biological Journal of the Linnean Society 103: 475–494. doi: 10.1111/j.1095-8312.2011.01656.x. [DOI] [Google Scholar]
- Shepherd L, Simon C, Langton-Myers S, Morgan-Richards M.. 2022. Insights into Aotearoa New Zealand’s biogeographic history provided by the study of natural hybrid zones. Journal of the Royal Society of New Zealand 52: 1–20. doi: 10.1080/03036758.2022.2061020. [DOI] [Google Scholar]
- Šmarda P, Stančík D.. 2005. Ploidy level variability in South American fescues (Festuca L. Poaceae): use of flow cytometry in up to 5½-year-old caryopses and herbarium specimens. Plant Biology 8: 73–80. [DOI] [PubMed] [Google Scholar]
- Soliani C, Gallo L, Marchelli P.. 2012. Phylogeography of two hybridizing southern beeches (Nothofagus spp.) with different adaptive abilities. Tree Genetics and Genomes 8: 659–673. doi: 10.1007/s11295-011-0452-9. [DOI] [Google Scholar]
- Soliani C, Tsuda Y, Bagnoli F, Gallo LA, Vendramin GG, Marchelli P.. 2015. Halfway encounters: meeting points of colonization routes among the southern beeches Nothofagus pumilio and N. antarctica. Molecular Phylogenetics and Evolution 85: 197–207. doi: 10.1016/j.ympev.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Somlo R, Durañona G, Ortiz R.. 1985. Valor nutritivo de especies forrajeras patagónicas. Revista de Producción Animal 5: 589–605. [Google Scholar]
- Soreng RJ, Peterson PM, Romaschenko K, et al. 2017. A worldwide phylogenetic classification of the Poaceae (Gramineae) II: an update and a comparison of two 2015 classifications: phylogenetic classification of the grasses II. Journal of Systematics and Evolution 55: 259–290. doi: 10.1111/jse.12262. [DOI] [Google Scholar]
- Španiel S, Rešetnik I.. 2022. Plant phylogeography of the Balkan Peninsula: spatiotemporal patterns and processes. Plant Systematics and Evolution 308: 38. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J.. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gaudeul M, Debussche M.. 2010. Conservation value of sites of hybridization in peripheral populations of rare plant species. Biological Conservation 24: 236–245. doi: 10.1111/j.1523-1739.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- Torrecilla P, Catalán P.. 2002. Phylogeny of broad-leaved and fine-leaved Festuca lineages (Poaceae) based on nuclear ITS sequences. Systematic Botany 27: 241–251. doi: 10.1043/0363-6445-27.2.241. [DOI] [Google Scholar]
- Torrecilla P, López Rodríguez JA, Stancik D, Catalán P.. 2003. Systematics of Festuca L. sects. Eskia Willk., Pseudatropis Kriv., Amphigenes (Janka) Tzvel., Pseudoscariosa Kriv. and Scariosae Hack. based on analysis of morphological characters and DNA sequences. Plant Systematics and Evolution 239: 113–139. doi: 10.1007/s00606-002-0265-2. [DOI] [Google Scholar]
- Tremetsberger K, Urtubey E, Terrab A, et al. 2009. Pleistocene refugia and polytopic replacement of diploids by tetraploids in the Patagonian and Subantarctic plant Hypochaeris incana (Asteraceae, Cichorieae). Molecular Ecology 18: 3668–3682. doi: 10.1111/j.1365-294X.2009.04298.x. [DOI] [PubMed] [Google Scholar]
- Vera-Escalona I, D’Elía G, Gouin N, et al. 2012. Lizards on ice: evidence for multiple refugia in Liolaemus pictus (Liolaemidae) during the Last Glacial Maximum in the southern Andean beech forests. PLoS One 7: e48358. doi: 10.1371/journal.pone.0048358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba R, Lara A, Boninsegna JA, et al. 2003. Large-scale temperature changes across the southern Andes: 20th-century variations in the context of the past 400 years. In: Diaz HF, ed. Climate variability and change in high elevation regions: past, present and future. Dordrecht: Springer, 177–232. doi: 10.1007/978-94-015-1252-7_10. [DOI] [Google Scholar]
- Wei N, Cronn R, Liston A, Ashman T.. 2019. Functional trait divergence and trait plasticity confer polyploid advantage in heterogeneous environments. New Phytologist 221: 2286–2297. doi: 10.1111/nph.15508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S, Keeling J, Gillman L.. 2006. The road from Santa Rosalia: a faster tempo of evolution in tropical climates. Proceedings of the National Academy of Sciences of the USA 103: 7718–7722. doi: 10.1073/pnas.0510383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.