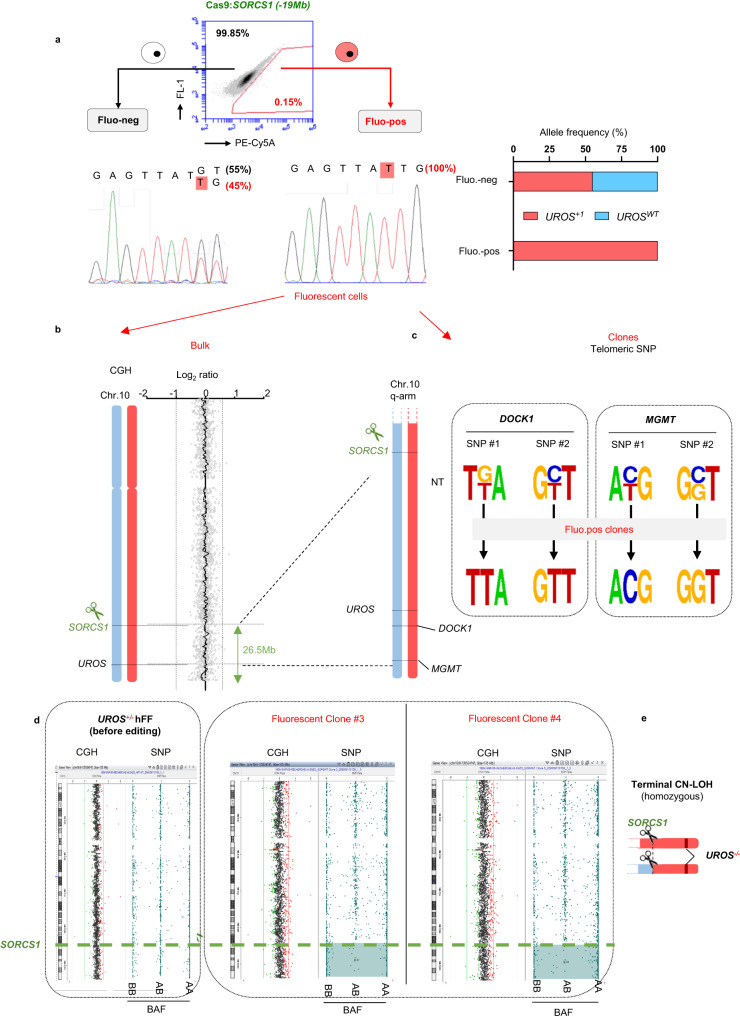
Fig. 3. hFAMReD reveals large rearrangements in hFFs after SORCS1 editing.
a After Cas9:SORCS1 editing, fluorescent-positive and negative cell sorting for UROS Sanger sequencing and LOH confirmation. b Array cGH of fluorescent-positive cell bulk showing no copy number variation. c SNP Sanger sequencing of DOCK1 (located at +1.4 Mb from UROS) and MGMT (+3.4 Mb) in non-transfected (NT) cells and in fluorescent-positive clones. d Array CGH and SNP array of cells before editing and two fluorescent-positive clones. Log2ratio and BAF (B-allele frequency) in control-hFFs (before editing) and two edited fluorescent clones (#3 and #4). Regions with loss of heterozygous AB alleles in blue. e Illustration of megabase-scale rearrangement detected by hFAMReD in hFFs after SORCS1 editing (CN-LOH). Source data are provided as a Source Data file.