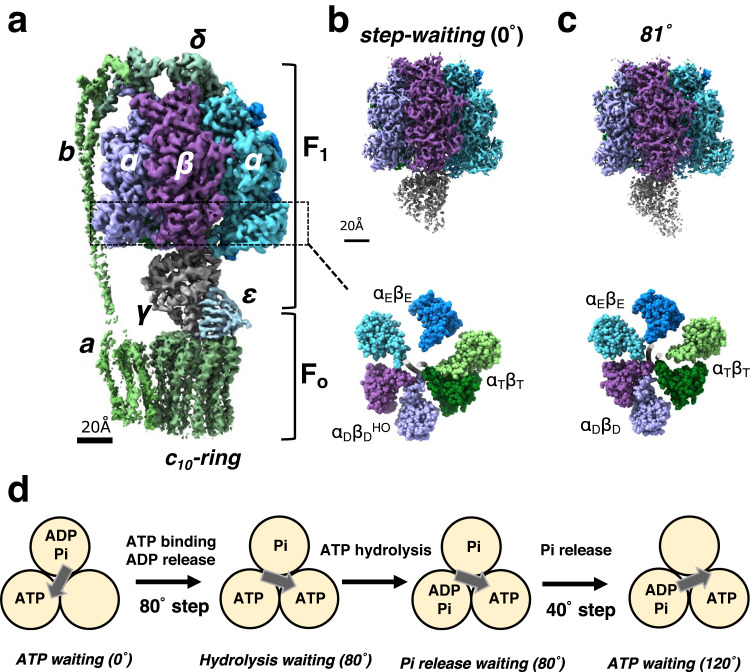
Fig. 1. Structures and rotation mechanism of FoF1.
a Cryo-EM structure of FoF1 ATP synthase in the step-waiting conformation. Each subunit is colored differently. The F1 domain contains three catalytic αβ dimers which surround the γ subunit. b, c Side views (upper) and bottom section views (lower) of the F1 domain in the step-waiting (0°) and 81° structure, respectively. The three catalytic dimers are represented different colors: αE (marine blue) and βE (light blue), αT (moss green) and βT (light green), and αD (purple) and βD (light purple). The βD subunit in step-waiting adopts a more open structure, termed as DHO. The γ subunits are represented as a gray tube in the center of both α3β3 sub-complexes. d A proposed scheme for chemo-mechanical coupling during a 120° rotation step of the F1 domain driven by ATP. In this model, ATP binding to the F1 domain immediately initiates the 80° rotation with an associated release of ADP. ATP is hydrolyzed at the 80° dwell position with no associated rotation of γ. It is the release of Pi from the enzyme which is suggested to drive the final 40° rotation.