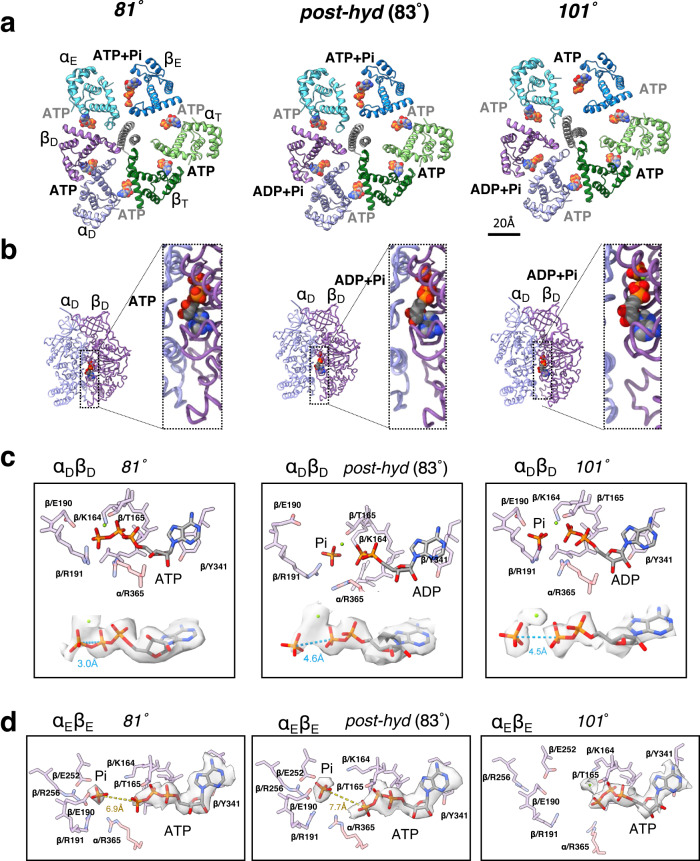
Fig. 2. Structure of 81° (left), post-hyd (center), and 101°(right) at high [ATP].
a Cross section of the F1 domain showing the catalytic sites viewed from the Fo side. Each catalytic dimer is shown in ribbon representation and colored as detailed in Fig. 1. The bound nucleotides are represented as spheres. b Side view structure of αDβD dimer. The left panel is a magnified view of the catalytic interface at each αDβD. c Structure of the catalytic site in αDβD. Amino acid residues and bound nucleotide and Pi are represented as sticks. Lower panels; Stick representation model of ATP/ADP and Pi with EM density in αDβD. The distance between γ and β phosphate of the ATP in each conformational state is shown in blue (Å). Each distance was calculated using Chimera software. d Structure of the catalytic site in αEβE. The EM density of ATP / Pi is superimposed onto the model. The distance between Pi and γ phosphate of ATP is shown in yellow (Å).