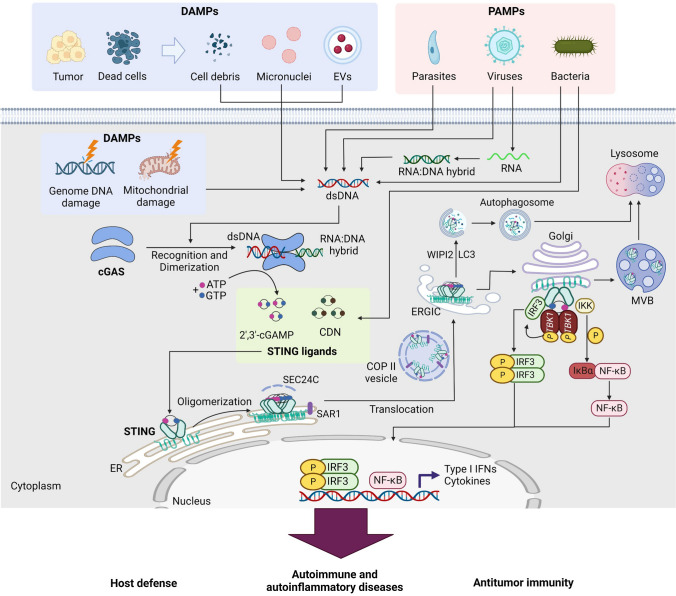
Fig. 1.
Activation of the cGAS/STING pathway by microbial DNA and self-DNA. Double-stranded DNA (dsDNA) derived from various sources of damage-associated molecular patterns (DAMPs), such as damaged cells and cancer cells, as well as dsDNA from various pathogen-associated molecular patterns (PAMPs) and RNA:DNA hybrids, activate the enzyme cyclic GMP-AMP synthase (cGAS) to synthesize 2′,3′-cyclic GMP-AMP (cGAMP), which serves as a STING ligand. Additionally, bacterial-derived cyclic dinucleotides (CDNs) act as STING ligands. Upon binding of STING ligands, STING translocates from the endoplasmic reticulum (ER) to the ER-Golgi intermediate compartment (ERGIC) via a process triggered by STING oligomerization and dependent on the SAR1 and SEC24C components. Within ERGIC, cGAMP-bound STING serves as a membrane source for the recruitment and lipidation of LC3 through a mechanism that is dependent on WIPI2. The resulting LC3-positive membranes then target DNA and pathogens to autophagosomes, which subsequently fuse with lysosomes. During translocation from the ERGIC to the Golgi, STING recruits TANK-binding kinase 1 (TBK1) and IκB kinase (IKK). This leads to phosphorylation of IRF3, which dimerizes and translocates to the nucleus to activate transcription of genes encoding type I interferons, including interferon-β (IFN-β). Phosphorylation of IκBα translocates NF-κB to the nucleus, where it activates the transcription of genes encoding proinflammatory cytokines, such as IL-6 and tumor necrosis factor (TNF). Finally, cGAMP-bound STING can also translocate to lysosomes for degradation via the multivesicular body (MVB) pathway, which involves the Golgi and endosomes