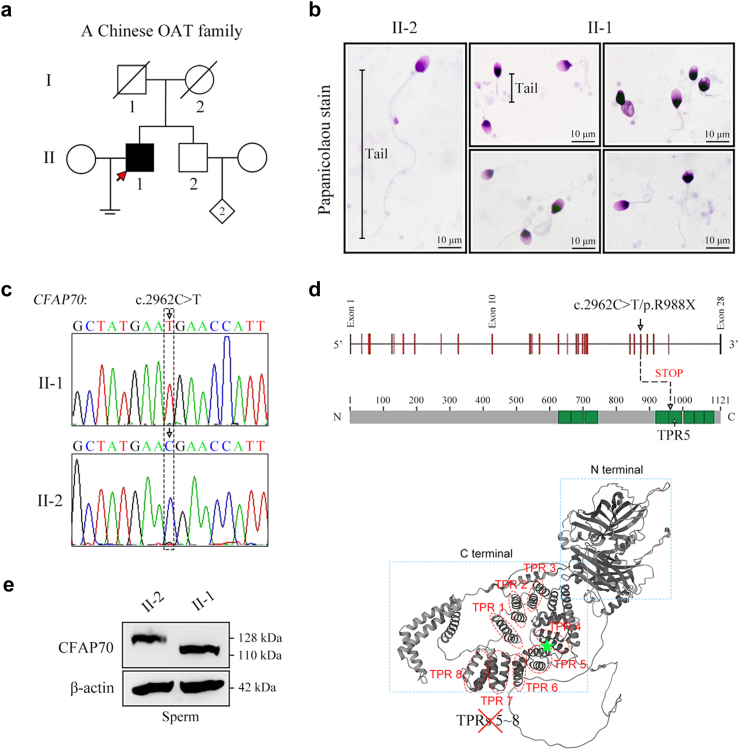
Fig. 5.
OAT phenotype of a CFAP70-mutated patient from a Chinese family. (a) Identification of a homozygous nonsense variant, c.2962C > T/p.R988X, of the CFAP70 gene in an OAT patient (II-1). His brother (II-2) is fertile. The pedigree of this family is presented, and the proband (II-1) is indicated by the arrow. (b) Representative sperm morphology in the proband (II-1) and his brother (II-2) by Papanicolaou staining under light microscopy. The spermatozoa of the proband (II-1) exhibited multiple flagellar malformations, including short, coiled, and absent flagella. Scale bars, 10 μm. (c) The DNA samples of the proband (II-1) and his brother (II-2) were subjected to Sanger sequencing. The chromatogram of CFAP70 in the proband (homozygous c.2962C > T) and his brother (wild-type allele). The arrow denotes the variant site. (d) The location of the variant (c.2962C > T/p.R988X) in the intron‒exon structure of the CFAP70 gene and the protein domain map of CFAP70. Eight tetratricopeptide repeats (TPRs) are annotated with green squares. The variant was located in the fifth TPR domain of CFAP70 and is expected to generate a truncated CFAP70 protein lacking 5–8 TPRs. The structure of CFAP70 was predicted by AlphaFold (https://alphafold.ebi.ac.uk/), and eight TPRs were shown. (e) The proteins of sperm samples were extracted from the proband (II-1) and his brother (II-2). Western blot analysis indicated a full-length CFAP70 protein (128 kDa) in II-2 and a truncated CFAP70 protein (∼110 kDa) in II-1. β-actin served as a loading control. Western blotting experiments were repeated for three times and representative blots were shown.