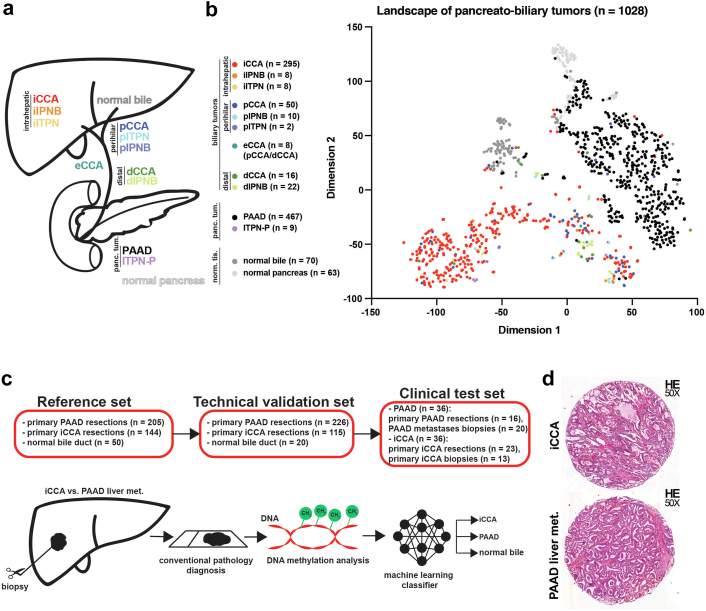
Fig. 1.
The methylation landscape of pancreato-biliary tumours. (a) Graphical representation of the pancreato-biliary tumours and normal tissues represented in the t-SNE plot. (b) The two-dimensional representation of pancreato-biliary tumours analysed using the t-SNE method based on DNA methylation profiles. The colour of the samples represents the tumour type and their origin: iCCA—intrahepatic cholangiocarcinoma; iIPNB—intrahepatic intraductal papillary neoplasia of the bile duct; iITPN—intrahepatic intraductal tubulopapillary neoplasia of the bile duct; pCCA—perihilar cholangiocarcinoma; pIPNB—perihilar intraductal papillary neoplasia of the bile duct; pITPN—perihilar intraductal tubulopapillary neoplasia of the bile duct; eCCA—extrahepatic cholangiocarcinoma (samples that are pCCA/dCCA but no further details were available); dCCA—distal cholangiocarcinoma; dIPNB—distal intraductal papillary neoplasia of the bile duct; PAAD—pancreatic adenocarcinoma; ITPN-P–intraductal tubulopapillary neoplasia of the pancreas; normal bile; and normal pancreas. (c) The design of the methylation-based classifier that can distinguish between iCCA, PAAD liver metastases and normal bile duct samples. (d) Representative H&E images for an iCCA and a PAAD liver metastasis.