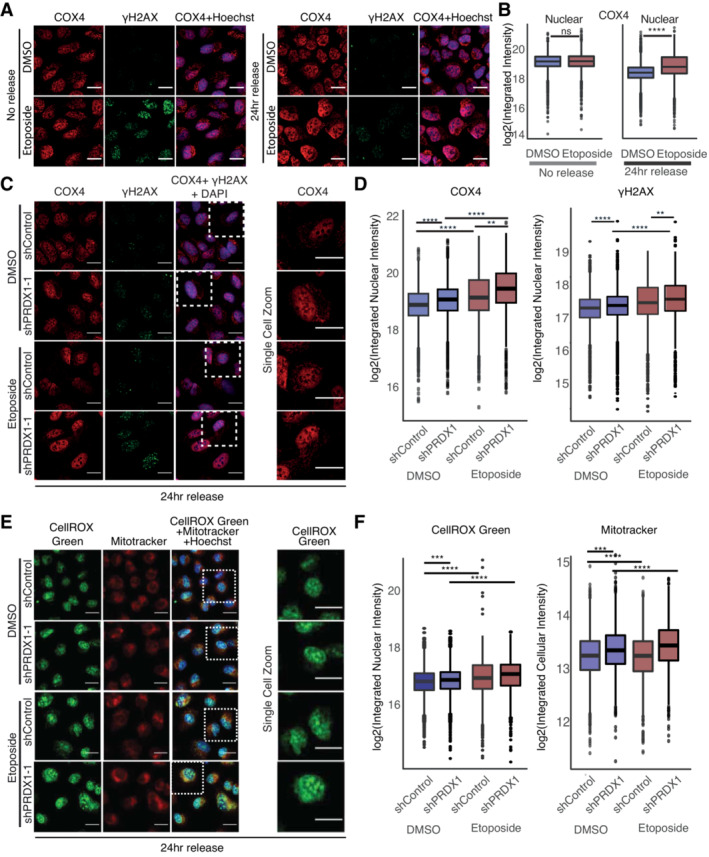
Visualization of COX4 (in red) and γΗ2ΑΧ (in green) within Hoechst stained nuclei (in blue) in U2‐OS WT cells at the indicated treatment conditions. Images were acquired with the Operetta High Content Screening System in confocal mode, scale bar is 25 μm.
Quantification of images shown in (A), represented as nuclear integrated intensities of COX4 signals. Three biological replicates were performed. A minimum of 1,000 cells were quantified for each condition, using Harmony. Boxplots represent the median with the IQR. P‐values were calculated using a Student's t‐test on the log2 normalized values (ns: not significant (P > 0.05), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Visualization of COX4 (in red) and γΗ2ΑΧ (in green) within DAPI stained nuclei (in blue) in U2‐OS shControl and shPRDX1 cells at the indicated treatment conditions. Images were acquired with the Operetta High Content Screening System in confocal mode, scale bar is 25 μm.
Quantification of images shown in (C), represented as nuclear integrated intensities of γΗ2ΑΧ and COX4 signals. Three biological replicates were performed. A minimum of 1,000 cells were quantified for each condition, using Harmony. Boxplots represent the median with the IQR. P‐values were calculated using linear regression on the log2 normalized values (ns: not significant (P > 0.05), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). The interaction term P‐value between PRDX1 background and etoposide treatment is shown in the plot.
Visualization of ROS (CellROX Green, in green) and mitochondria (Mitotracker, in red) within Hoechst‐stained nuclei (in blue) in U2‐OS shControl and shPRDX1 cells at the indicated treatment conditions. Images were acquired with the Operetta High Content Screening System in confocal mode, scale bar is 25 μm.
Quantification of images shown in (E), represented as log2 nuclear‐integrated intensity of CellROX Green and Mitotracker immediately at 24 h release compared to DMSO control. Three biological replicates were performed. A minimum of 1,000 cells were quantified for each condition, using Harmony. Boxplots represent the median with the IQR. P‐values were calculated using linear regression on the log2 normalized values (ns: not significant (P > 0.05), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). The interaction term P‐value between PRDX1 background and etoposide treatment is shown in the plot.