Abstract
In non-small cell lung cancer (NSCLC), the heterogeneity promotes drug resistance, and the restricted expression of programmed death-ligand 1 (PD-L1) limits the immunotherapy benefits. Based on the mechanisms related to translation regulation and the association with PD-L1 of methyltransferase-like 3 (METTL3), the novel small-molecule inhibitor STM2457 is assumed to be useful for the treatment of NSCLC. We evaluated the efficacy of STM2457 in vivo and in vitro and confirmed the effects of its inhibition on disease progression. Next, we explored the effect of STM2457 on METTL3 and revealed its effects on the inhibition of catalytic activity and upregulation of METTL3 protein expression. Importantly, we described the genome-wide characteristics of multiple omics data acquired from RNA sequencing, ribosome profiling, and methylated RNA immunoprecipitation sequencing data under STM2457 treatment or METTL3 knockout. We also constructed a model for the regulation of the translation of METTL3 and PD-L1. Finally, we found PD-L1 upregulation by STM2457 in vivo and in vitro. In conclusion, STM2457 is a potential novel suppressor based on its inhibitory effect on tumor progression and may be able to overcome the heterogeneity based on its impact on the translatome. Furthermore, it can improve the immunotherapy outcomes based on PD-L1 upregulation in NSCLC.
Keywords: STM2457, METTL3, Translatomics, Non-small cell lung cancer, PD-L1
Graphical abstract
Highlights
-
•
STM2457, the novel small-molecule inhibitor of METTL3, inhibits NSCLC progression.
-
•
STM2457 inhibits the catalytic activity of METTL3 and increases its protein level.
-
•
STM2457 improves the translation efficiency of genes such as PD-L1 and METTL3.
1. Introduction
Lung cancer is the leading cause of cancer-related death and the second most common cancer worldwide [1]. The majority of lung cancers (80%–85%) are non-small cell lung cancer (NSCLC), characterized by a group of distinct diseases with genetic and cellular heterogeneity [2]. This heterogeneity promotes drug resistance, which is the major reason for the failure of currently available therapeutic approaches [3]. In the last decade, the development of antibodies against programmed death-1 (PD-1) receptor and programmed death-ligand 1 (PD-L1) have led to a significantly prolonged survival for a large proportion of NSCLC patients. However, immunotherapies have limited activity due to the restricted expression of receptor antigens [4], and the five-year overall survival for NSCLC patients remains less than 20%. Therefore, there is an urgent need to develop novel molecules that prevent disease progression, overcome heterogeneity, and expand the patients eligible for immunotherapy in NSCLC.
Because the translation mechanism integrates almost all oncogenic signals and has extensive and crucial effects on oncogene protein expression, targeting the components of this machinery is promising for overcoming disease heterogeneity [5]. The newly revealed translation mechanisms by Choe et al. [6] have increased the interest in methyltransferase-like 3 (METTL3), the key enzyme involved in N6-methyladenosine (m6A) methylation. The mRNA circularization by METTL3 promotes the translation of oncogenes, such as epidermal growth factor receptor and Yes1-associated transcriptional regulator, in human cancer cells [6]. Furthermore, accumulating evidence indicates that METTL3 oncogene is significantly overexpressed and promotes disease progression in NSCLC, but the underlying mechanism remains unclear [7]. Previous studies of various cancers, including bladder cancer, have reported an association between METTL3 and PD-L1 expression through sustained mRNA stability, among other mechanisms [8]. As a result, METTL3 may be a novel therapeutic target for NSCLC. STM2457 is the first bioavailable inhibitor of METTL3 and may be a potential therapeutic strategy in NSCLC to inhibit disease progression, overcome heterogeneity, and extend the benefits of immunotherapy.
STM2457 was initially synthesized by Yankova et al. [9] and proved to be effective for the treatment of acute myeloid leukemia. In recent years, the antitumor effect of STM2457 in solid tumors such as intrahepatic cholangiocarcinoma [10] and Sonic hedgehog medulloblastoma [11] has been reported, but its effects and mechanism in NSCLC remain to be further explored. We evaluated the use of STM2457 in NSCLC in vivo and in vitro, explored its effect on METTL3, revealed the genome-wide characteristics of multiple omics data (especially translatomics), and explored the regulation for PD-L1. The results of the in vitro and in vivo effects of STM2457 enabled us to determine the potential use of the small-molecule inhibitor and the feasibility of targeting METTL3. The effects of the inhibitor on the molecular target METTL3 and translatomics characteristics will guide the development of targeted translation to overcome the heterogeneity in NSCLC. Finally, the evidence that STM2457 regulates PD-L1 expression will strongly support the use of a combination of STM2457 and anti-PD-l/PD-L1 to improve the immunotherapy benefits.
2. Material and methods
2.1. Cell culture
Human lung cancer cell lines (A549, H1975, H1793, and HCC827) and normal lung epithelial cell lines (HBE135-E6E7 and BEAS-2B) were obtained from the American Type Culture Collection (Manassas, VA, USA). Another lung cancer cell line PC-9 was purchased from the European Collection of Authenticated Cell Cultures (Salisbury, UK). Dulbecco's Modified Eagle Medium-high glucose (Thermo Fisher Scientific Inc., Waltham, MA, USA) medium supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin, and 1% streptomycin (Gibco, Carlsbad, CA, USA) was applied to maintain the cells at 37 °C with 5% CO2. A549 and H1975 were selected as representative NSCLC cell lines of the current study due to their capacity for high proliferation, high metastasis, and high sensitivity to STM2457, which benefitted us in exploring the effects and mechanism of STM2457 in vitro.
2.2. STM2457 treatment
STM2457 (MedChemExpress, Monmouth Junction, NJ, USA) was synthesized based on the published chemical structure. The molecular formula of STM2457 (CAS No.: 2499663-01-1) is C25H28N6O2 with a molecular weight of 444.53. STM2457 was transported in powder form and can be stored for three years at −20 °C and two years at 4 °C. In in vitro experiments, STM2457 was dissolved in dimethylsulfoxide (DMSO) to prepare a storage solution with a concentration of 10 mM, and then the storage solution was diluted with the complete medium to a series of working solutions with various concentrations as required. In in vivo experiments, STM2457 was dissolved in a 20% (m/V) 2-hydroxypropyl-beta-cyclodextrin (Glpbio, Montclair, CA, USA) vehicle as the working solution with a concentration of 5 mg/mL. After one month of modeling, 50 mg/kg of STM2457 or the respective vehicle was delivered to the mice via intraperitoneal (i.p.) injection once daily for a total of two weeks (14 treatments).
2.3. Cell viability assay
Based on the variety of morphology, proliferation, STM2457 reactivity, and treatment time, the aforementioned cell lines were inoculated into 96-well plates at different densities. To evaluate the viability of cells treated with 6-day STM2457, A549, PC-9, HBE, HCC827, and H1793 were inoculated at 2,000 cells/well, whereas H1975 and BEAS-2B were inoculated at 1,000 cells/well. Moreover, for the cell viability evaluation of 3- and 1-day STM2457 treatment, a slightly higher density of A549 (3,000 and 5,000 cells/well) and H1975 (2,000 and 5,000 cells/well) inoculation was necessary. After overnight inoculation, the complete medium was replaced with gradient concentrations (0, 0.01, 0.05, 0.1, 0.5, 1, 5, 10, and 50 μM) of working solution from 0 to 50 μM or vehicle (DMSO) for 1, 3, or 6 days. At the end of the STM2457 treatment, the supernatant was removed, and Cell Counting Kit-8 (Servicebio, Wuhan, Hubei, China) was evenly mixed with a complete medium at a ratio of 1:9 and then added into 96-well plates (100 μL/well). After incubation at 37 °C for 1 h, the absorbance at 450 nm was measured using a microplate reader (Thermo Fisher Scientific Inc.). All experiments were carried out in triplicate.
2.4. Colony formation assay
To evaluate the effect of STM2457 on A549 and H1975 proliferation, 1,000 cells were seeded per well in a six-well plate. The cells were cultured with STM2457 (0, 1, and 5 μM) and DMSO-containing medium for 10 days at 37 °C with 5% CO2 until the number of cells in a single clone was more than 50, and the medium was replaced every three days. At the end of observation, the cells were fixed with 4% paraformaldehyde and stained with crystal violet (Servicebio).
2.5. Wound-healing assay
A549 and H1975 were pretreated with 5 μM of STM2457 and vehicle (DMSO) for 1, 3, and 6 days, and ibidi Culture-Insert 2 Well in μ-Dish 35 mm (ibidi, Fitchburg, WI, USA) was applied for wound-healing assay to evaluate the migration ability of A549 and H1975. Referring to the protocol, 50,000 cells were suspended with 70 μL of complete medium and seeded equally into two wells of culture-insert in a 35-mm dish. The cells were incubated overnight at 37 °C with 5% CO2 to ensure adherence and 90% confluence, the culture insert was extracted, and a cell-free gap with a width of 500 ± 100 μm was formed between cell populations in two wells. The original complete medium was replaced by low serum (2% FBS) medium containing 5 μM of STM2457 or DMSO consistent with pretreatment condition. During 24 h of culture, the changes in cell-free gaps in each pretreatment group were observed and photographed regularly (0, 3, 6, 12, and 24 h). Each assay was performed at least three times.
2.6. Matrigel transwell assay
Matrigel transwell assay was conducted using a 6.5-mm transwell with 8.0-μm pore polycarbonate membrane inserts (Corning Inc., Corning, NY, USA) coated with BD Matrigel basement membrane matrix (BD Biosciences, Franklin Lakes, NJ, USA) to evaluate the invasion ability of A549 and H1975 pretreated with 5 μM of STM2457 for 1, 3, and 6 days or DMSO for 6 days. In the invasion assay, 50,000 cells suspended in 200 μL of serum-free medium were seeded in the upper compartment of the chamber, and 800 μL of Dulbecco's Modified Eagle Medium with 20% FBS was added to the lower compartment of the chamber. STM2457 and DMSO were mixed evenly with the aforementioned medium to maintain the pretreatment conditions, and the cells were incubated for another 24 h. After that, the cells were fixed with 4% paraformaldehyde for 15 min and stained with crystal violet. The non-invading cells in the upper chamber were removed, and the invaded cells on the lower surface were photographed in five randomly selected visual fields. Each assay was performed at least three times.
2.7. Western blot
Total proteins in NSCLC cell lines were extracted with radioimmune precipitation assay buffer (Servicebio), quantified by a bicinchoninic acid protein assay kit (Servicebio), separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, and then transferred into polyvinylidene fluoride membranes. The membranes were incubated overnight with primary antibody against METTL3 (ab195352; Abcam, Cambridge, UK), PD-L1 (ab205921; Abcam), poly(adenosine diphosphate-ribose) polymerase (PARP) (#9532; Cell Signaling Technology, Inc., Danvers, MA, USA), caspase-3 ( #9664; Cell Signaling Technology, Inc.), B-cell lymphoma-2 (Bcl-2) (#15071; Cell Signaling Technology, Inc.), Bcl-2-associated X protein (Bax) (#41162; Cell Signaling Technology, Inc.), and glyceraldehyde-3-phosphate dehydrogenase (ab8245; Abcam) at 4 °C. After washing, horseradish peroxidase (HRP)–conjugated secondary antibodies (SA00001-1 and SA00001-2; Proteintech Group, Inc., Rosemont, IL, USA) were applied to incubate membranes for 2 h at room temperature, and then the blot signals were detected using an enhanced chemiluminescence (ECL) detection system (ChemiDoc XRS+ machine; Bio-Rad Laboratories, Hercules, CA, USA).
2.8. Apoptosis assay
Cell apoptosis was evaluated based on the fluorescence-activated cell sorting analysis by Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining. A549 and H1975 treated with STM2457 (0, 1, and 5 μM) and DMSO for 3 or 6 days were harvested, washed twice in ice-cold phosphate-buffered saline (PBS) with 2% bovine serum albumin, and centrifuged at 2,000 rpm for 5 min to collect 1 × 105–5 × 105 cells. The cells were resuspended in 500 μL of binding buffer and stained for 5–10 min with 5 μL of Annexin V-FITC and 5 μL of PI using an Annexin V-FITC/PI staining kit (Jiangsu KeyGene BioTech Corp., Ltd., Nanjing, China). The status of cell apoptosis was analyzed by BD AccuriTM C6 Plus flow cytometer (BD Biosciences).
2.9. Animal
A total of six female BALB/c nude mice (Shulb, Wuhan, Hubei, China) aged 4–6 weeks were raised under specific pathogen-free conditions with a temperature of 22 ± 2 °C and humidity of 55% ± 5% on a 12-h light/dark cycle. They were allowed ad libitum access to food and water during the experiments. All of the animal experiments were performed according to the guidelines of the Institutional Animal Care and Use Committee of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, Hubei, China), which also approved the protocols (Approval No.: 2847).
2.10. Mouse model construction and administration
After 2 weeks of acclimatization, six female BALB/c nude mice aged 6–8 weeks were injected with A549 suspension (1 × 106) on the tail vein to construct a pulmonary metastasis mouse model and randomly divided into two groups. After 4 weeks of modeling, 50 mg/kg of STM2457 and the respective vehicle (20% 2-hydroxypropyl-beta-cyclodextrin) were delivered to the mice via i.p. injection once daily for a total of 2 weeks (14 treatments). A total of 6 weeks later, mice were sacrificed to observe pulmonary foci and detect the expression of targets.
2.11. m6A modification level quantification
According to the protocol of EpiQuik m6A RNA methylation Quantification Kit (Colorimetric) (EpigenTek, Farmingdale, NY, USA), the m6A RNA methylation status of total RNA isolated from A549 and H1975 under STM2457 treatment was directly detected, which indirectly reflected the effect of STM2457 on the catalytic activity of METTL3. Total RNA was extracted from the aforementioned cells with TRIzol reagent (Thermo Fisher Scientific Inc.) and 200 ng of sample RNA (1–8 μL) was bound into the assay wells. Meanwhile, negative control reagents and diluted positive control reagents were used for the preparation of the standard curve. Capture antibody, detection antibody, and enhancer solution were successively added to capture RNA modified with m6A methylation. Each well was incubated with developer solution at room temperature for 1–10 min away from light, the enzyme reaction was stopped after adding stop solution, and the absorbance was read on a microplate reader (PerkinElmer Inc., Waltham, MA, USA) at 450 nm within 2–15 min. The standard curve using linear regression was drawn for absolute quantification of m6A methylated RNA. Measurements were performed in triplicate.
2.12. RNA sequencing
Total RNA from 1 × 107 cells was extracted using TRIzol reagent for RNA sequencing (RNA-seq). VAHTS Stranded mRNA-seq Library Prep Kit for Illumina V2 (Vazyme Biotech, Nanjing, Jiangsu, China) was used for library preparation according to the instructions. Reads were aligned to the human Ensemble genome GRCh38 using Hisat2 aligner (v2.1.0) under parameters: -rna-strandness RF. The reads mapped to the genome were calculated using featureCounts (v1.6.3). Differential gene expression analysis was performed using the DESeq2 R-package.
2.13. Ribo sequencing
A total of 1 × 107 cells were prepared for cell lysis. Then, the cell medium was discarded, and the cells were washed twice with ice-cold PBS containing 100 μg/mL of cycloheximide (Adooq Bioscience, Irvine, CA, USA). Ribosome profiling was performed using Epi Ribosome Profiling Kit (Epibiotek, Guangzhou, China). Subsequently, ribosome-protected RNA fragments were extracted using an RNA Clean & Concentrator-5 kit (Zymo Research, Orange County, CA, USA). Epi RiboRNA Depletion Kit (Human/Mouse/Rat) (Epibiotek) was used for rRNA depletion. Sequencing libraries were constructed using the QIAseq miRNA Library kit (QIAGEN, Dusseldorf, Germany). Adapter sequences were removed from raw sequencing data using cutadapt software. Meanwhile, reads with a length between 25 and 35 bp were kept for downstream analysis. Then, reads were aligned to rRNA and tRNA sequences to remove rRNA and tRNA reads using bowtie software, and the remaining reads were used to align to reference genome and transcriptome (Ensembl Version 91) using hisat2 and bowtie software separately. The trinucleotide periodicity of ribosomes and codon usage frequency were estimated using the revised riboWaltz package. Open reading frame identification was performed using Price software. Read counts were calculated using featureCounts software. Raw counts were further normalized as reads per kilobase per million mapped reads values using the fpkm function in the DESeq2 package. Translation efficiencies (TEs) were determined as the ratio of (normalized abundance determined by ribosome profiling)/(normalized abundance determined by RNA-seq), as previously reported [12].
2.14. Methylated RNA immunoprecipitation sequencing (meRIP-seq)
Total RNA from 1 × 107 cells was extracted using TRIzol reagent, and DNA contamination was removed using DNase I (BBI Solutions, Cardiff, Wales, UK). Next, the precipitated RNA was washed with ethanol and dissolved in ultrapure water. After concentration determination, RNA was shared into approximately 200-nt fragments by incubation at 70 °C for 7 min in a water bath. Of the aforementioned fragmented RNA, 1/10 was saved as the input control for further RNA-seq, and the remaining RNA was incubated with m6A (D9D9W) Rabbit mAb (Cell Signaling Technology, Inc.) for 2 h at 4 °C and then mixed with DynabeadsTM Protein A for Immunoprecipitation (Thermo Fisher Scientific Inc.) at 4 °C for 2 h to acquire methylated RNA complex. The m6A antibody was digested with Proteinase K digestion buffer (TransGen Biotech, Beijing, China), and the methylated RNA was purified for further meRIP-seq by DIATRE (Shanghai, China).
2.15. Bioinformatics analysis
The upstream processing of raw data acquired from the aforementioned multiple omics sequencing, such as quality control (Fastqc), alignment (Hisat2, bowtie, and STAR), peak calling (MACS2), and annotation (deeptools), was based on Python (version 3.9.12) and Java (version 1.8.0) of Linux. The statistical analysis and visualization of clean data were mainly performed based on R (version 4.2.1) of Windows. The results of the differential gene expression analysis were visualized in the forms of volcano, dumbbell, and scatter plots performed using the ggplot2/R package. The clusterProfiler package was used for Gene Ontology (GO)/Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. The Venn plots drawn using the VennDiagram package reflected the results of cross-analysis among differential expression genes in multiple omics sequencing. The results of motif analysis were presented through the R package ggseqlogo, the peak distribution fraction was depicted as the pie chart through the R package ggplot2, and the peak distribution profile was performed using plotProfile of deeptools. The software Integrative Genomics Viewer on Windows is a powerful tool for visualizing the results of reads mapped to the targets (METTL3 and PD-L1).
2.16. Interferon-gamma (IFN-γ) induction
A549 and H1975 were seeded into six-well plates and treated under different concentrations of STM2457 and/or 10 ng/mL of IFN-γ (MedChemExpress, Monmouth Junction, NJ, USA) for 24 h. After washing with PBS, total cells were obtained and recognized with primary and fluorescent antibodies: PD-L1 and APC anti-human CD274 (BioLegend, San Diego, CA, USA). Under the induction of IFN-γ, Western blot and flow cytometry assay were performed and analyzed for total and surface expression of PD-L1, respectively.
2.17. Flow cytometry assay
Harvested NSCLC cells were washed with PBS and incubated at 4 °C for 45 min away from light using 2% FBS and appropriate antibodies in PBS. After washing with PBS, the samples were detected on BD AccuriTM C6 Plus flow cytometer, and the data were analyzed in FlowJo 10.6.2 (Ashland, OR, USA).
2.18. CRISPR-Cas9
To knock out the METTL3 gene in A549 (A549 KO_METTL3), oligos for gRNAs were cloned into a lentiGuide-Puro vector (Addgene 52963) not containing Cas9 and transfected into A549 that stably expressed Cas9 by lentivirus infection. Three pairs of alternative gRNAs for the METTL3 gene were constructed with the help of Genomeditech (Shanghai, China). The following gRNAs were used for knocking out METTL3 genes in A549: METTL3 gRNA1 (gRNA1 5′: GGGCTGTCACTACGGAAGGT, gRNA1 3′: AGCATCAGTGGGCAATGTTA), METTL3 gRNA2 (gRNA2 5′: GATGCTCCTGCCACTCAAGA, gRNA2 3′: TGCTGCCTCAGATGTTGATC), and METTL3 gRNA3 (gRNA3 5′: GCTTTCTACCCCATCTTGAG, gRNA3 3′: GAGTTGATTGAGGTAAAGCG). Based on the verification of the gene expression effect and effective target, gRNA2 was selected from the aforementioned three pairs of gRNAs to continue the downstream experiment. After monoclonal preparation, culture, and a series of validation, the stable cell line with METTL3 knockout (A549 KO_METTL3) was finally obtained from A549 cells infected with H_METTL3 link-sgRNA2.
2.19. Dot blot of m6A modification level
mRNAs isolated from the total RNA of A549 KO_NC and KO_METTL3 cell lines treated with 5 μM of STM2457 and DMSO for 6 days were used to determine the concentration using the NanoDrop and diluted to 100, 50, and 25 ng/μL using ribonucleases-free water. The serially diluted mRNA was denatured at 95 °C for 3 min and chilled on ice immediately, and 2 μL of mRNA was dropped onto the positively charged nylon membranes (Beyotime, Shanghai, China). Spotted mRNA was crosslinked twice to the membrane in a Stratalinker 2400 UV Crosslinker (Analytik Jena US LLC, Upland, CA, USA) using the autocrosslink mode (1,200 μJ (×100); 25–50 s). After blocking, the membranes were incubated overnight with anti-m6A antibody (ABclonal, A19841; ABclonal, Wuhan, China) at 4 °C. Then, the HRP-conjugated Affinipure Goat Anti-Rabbit secondary antibody (SA00001-2; Proteintech Group, Inc.) was added to the blots for 1 h at room temperature, and the membrane was developed with the ECL detection system (ChemiDoc XRS+ machine).
2.20. METTL3-mut plasmid
Human METTL3 (NM_019852) and METTL3 catalytic mutant (aa395-398, DPPW/APPA) cDNAs were amplified by ClonExpress II One Step Cloning Kit (Vazyme) and then subcloned into the vector CMV-MCS-3FLAG-IRES-EGFP-SV40-Neomycin (GeneChem, Shanghai, China) to generate the plasmids of wild type (OE_METTL3) and catalytic defect METTL3 (OE_METTL3_Mut) overexpression.
2.21. Transfection of plasmid
Cells at 70%–90% confluence were transfected with plasmids using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) diluted in Opti-MEM Reduced Serum Medium (Gibco). After 2–4 days of incubation at 37 °C, the cells were collected for subsequent experiments.
2.22. Hematoxylin and eosin (H&E) staining
The whole lung specimens from the pulmonary metastasis mouse model were paraffin-embedded and sliced into tissue sections with a thickness of 3–4 μm. After dewaxing, the tissue sections were stained with hematoxylin dye (Servicebio) for 3–8 min, differentiated with 1% hydrochloric acid alcohol, and turned blue with 0.6% aqueous ammonia to complete nuclear staining, and cytoplasmic staining was performed by immersing sections in eosin dye (Servicebio) for 1–3 min. After dehydration and drying, the tissue sections were sealed with gum, digitally scanned, and analyzed.
2.23. Immunohistochemistry staining
Following the protocol of the immunohistochemistry secondary antibody kit (Absin, Shanghai, China), dewaxed and hydrated tissue sections were incubated with hydrogen peroxide for 10 min, blocking buffer for 5 min, and primary antibody against human METTL3 and human PD-L1 for 20 min at room temperature. After 10-min incubation of primary antibody amplifier and HRP-conjugated secondary antibody polymer, the fresh substrate solution was dropped on tissue sections and reacted for 5 min. After dehydration and drying, the tissue sections were sealed with gum, digitally scanned, and analyzed.
2.24. Statistical analysis
All analyses of results from molecular biology experiments, especially cell viability assay and m6A modification level detection, were performed using GraphPad Prism version 9.0 and OriginPro 2022. All data and error bars are presented as the mean ± standard deviations from at least three independent experiments. All differences between the two independent groups were evaluated by a two-tailed Student's t-test. The |log2fold change (FC)| > 1 and P < 0.05 were used as the criteria to define differential and stable expression in all differential analyses. The indicated P values (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001) were considered to be statistically significant.
3. Results and discussion
3.1. In vitro and in vivo effects of STM2457 on NSCLC progression
STM2457 (HY-134836) was synthesized based on the published chemical structure (Fig. 1A). Considering the initial application of STM2457 in NSCLC, the dose-response curves for 1, 3, and 6 days in A549 (Fig. 1B) and H1975 (Fig. 1C) cells were plotted to determine the optimal treatment time (6 days) and concentration (5 μM). Additionally, the half-maximal inhibitory concentration values were calculated (Fig. 1D). NSCLC cell lines (A549: 4.101 μM; H1975: 8.343 μM; PC-9: 3.758 μM; H1793: 11.66 μM; and HCC827: 14.59 μM) were more sensitive to STM2457 than normal lung epithelial cell lines (BEAS-2B: 22.187 μM and HBE: 20.21 μM).
Fig. 1.
The structure of STM2457 and its dose-response curves in non-small cell lung cancer (NSCLC). (A) Chemical structure of STM2457 (CAS# 2499663-01-1). Dose-response curves of STM2457 treatment at 1, 3, and 6 days in (B) A549 and (C) H1975 cells. (D) Dose-response curves of STM2457 treatment for 6 days in NSCLC cell lines (PC-9, A549, H1975, H1793, and HCC827) and normal lung epithelial cell lines (HBE and BEAS-2B).
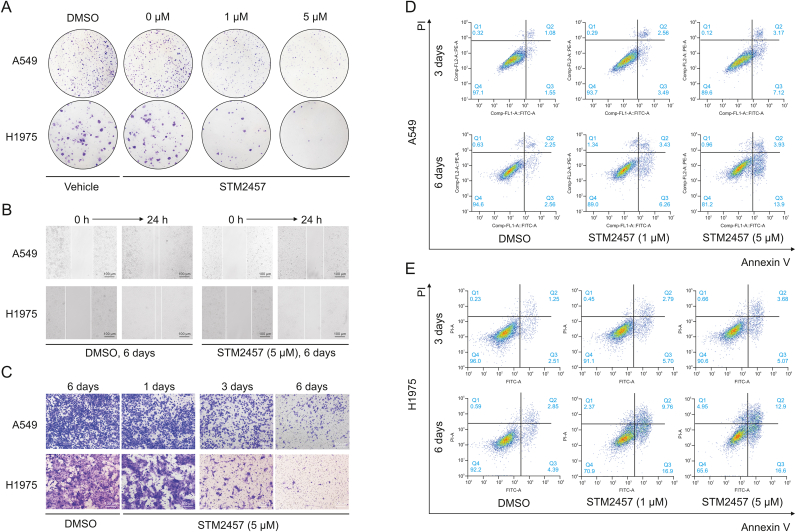
The in vitro effects of STM2457 on A549 and H1975 cells showed degenerated proliferation at 1 μM; the effect was particularly conspicuous at 5 μM in the colony-forming assay (Fig. 2A). Furthermore, treatment with 5 μM STM2457 for 6 days inhibited the cell migration ability by wound-healing assay (Figs. 2B and S1). The results of matrigel transwell assays suggested that the invasion capability of NSCLC cell lines was weakened by prolonged treatment with 5 μM STM2457 (Fig. 2C). Moreover, fluorescence-activated cell sorting analysis by Annexin V-FITC/PI staining revealed the apoptosis-inducing effect of STM2457 in A549 (Fig. 2D) and H1975 (Fig. 2E), which was supported by the consistent changes in apoptosis-related proteins (Fig. S2A and B) in which cleaved PARP (Figs. S2C and D) and caspase-3 (Figs. S2E and F) were hyperexpressed, and the ratio of Bcl-2 to Bax (Figs. S2G and H) was significantly decreased. As regards the administration regimen in acute myeloid leukemia, 50 mg/kg of STM2457 or the respective vehicle was delivered to the pulmonary metastasis mouse models daily via i.p. injection for 14 days (Fig. 3A). The results of H&E staining confirmed the inhibitory effect of STM2457 on pulmonary colonization and progression of NSCLC in vivo (Fig. 3B).
Fig. 2.
In vitro effects of STM2457 on non-small cell lung cancer (NSCLC) progression. (A) The effect of STM2457 on NSCLC proliferation was assessed using colony-forming assay in A549 and H1975 cells treated with STM2457 (0, 1, and 5 μM) or the respective vehicle (dimethylsulfoxide (DMSO), 5 μM) for 10 days. (B) The effect of STM2457 on NSCLC migration was evaluated using wound-healing assay for 24 h in A549 and H1975 cells pretreated with STM2457 (5 μM) or the respective vehicle DMSO for 6 days. (C) The effect of STM2457 on NSCLC invasion was verified using matrigel transwell assays for 48 h in A549 and H1975 cells pretreated with STM2457 (5 μM) for 1, 3, and 6 days or the respective vehicle DMSO for 6 days. The effect of STM2457 on NSCLC apoptosis was revealed using the fluorescence-activated cell sorting analysis by Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining in A549 (D) and H1975 (E) cells treated with STM2457 (1 and 5 μM) or the respective vehicle DMSO for 3 or 6 days.
Fig. 3.
In vivo effects and regulation of STM2457 on programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer (NSCLC). (A) The modeling and drug administration scheme of a pulmonary metastasis mouse model. (B) In the lung sections of a pulmonary metastasis mouse model, the effect of STM2457 on NSCLC progression and the regulation of STM2457 on the expression of both methyltransferase-like 3 (METTL3) and PD-L1 in vivo were evaluated using hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC) staining, respectively. HP: hydroxypropyl.
3.2. Molecular mechanism of effects of STM2457 in NSCLC
3.2.1. Comprehensive evaluation of the effect of STM2457 on METTL3 in NSCLC
To reveal the molecular mechanism underlying the aforementioned effects, we evaluated the effect of STM2457 on METTL3 in NSCLC. As an inhibitor of METTL3 catalytic activity, STM2457 upregulated the METTL3 protein expression in NSCLC in a time- and concentration- dependent manner (Figs. 4A and B). Similar to the MET receptor tyrosine kinase inhibitors [13], compensatory protein overexpression and protein accumulation are not rare with treatment with small-molecule inhibitors, suggesting the complex regulatory effects on METTL3. Therefore, STM2457 should be carefully considered as a potential treatment for NSCLC. Perhaps influenced by this compensatory mechanism, the activity of METTL3-mediated m6A RNA methylation showed a consistent inhibition and then activation with STM2457 treatment in a time- and concentration-dependent manner (Figs. 4C and D). The total cellular m6A in mRNA level was most significantly inhibited at day 3 (A549: P < 0.0005) or 4 (H1975: P < 0.0001) and at a concentration of 5 μM (A549: P < 0.0001 and H1975: P < 0.0001). Using the dose-response curves of STM2457 (Figs. 1B and C), we found that the optimum inhibitory effect of the drug at the level of m6A was earlier than the optimum inhibitory effect on cell proliferation. The weak effect relative to the inhibitors of oncogenic drivers, a common feature of epigenetic therapies including STM2457 [14], appears insufficient to explain the contradiction between the effect and the mechanism. However, whether the cytotoxicity of STM2457 is due to its role in other mechanisms than m6A modification needs to be further explored.
Fig. 4.
Impact of STM2457 on methyltransferase-like 3 (METTL3) expression and activity of non-small cell lung cancer (NSCLC). (A) The time-dependent effect of STM2457 on METTL3 expression was detected by Western blot in A549 and H1975 cells treated with STM2457 (5 μM) for 0–6 days or the respective vehicle (dimethylsulfoxide (DMSO), 5 μM) for 6 days. (B) The concentration-dependent effect of STM2457 for 3 or 6 days on METTL3 expression was detected by Western blot in A549 and H1975 cells treated with STM2457 (0, 0.01, 0.05, 0.1, 0.5, 1, 5, 10, and 50 μM) or the respective vehicle (DMSO, 5 μM). (C) The time-dependent effect of STM2457 on the activity of METTL3-mediated N6-methyladenosine (m6A) RNA methylation modification was evaluated using EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) in A549 and H1975 cells treated with STM2457 (5 μM) for 0–6 days or the respective vehicle (DMSO, 5 μM) for 6 days. (D) The concentration-dependent effect of STM2457 on METTL3 activity was evaluated in A549 and H1975 cells treated with STM2457 (0, 0.01, 0.05, 0.1, 0.5, 1, 5, 10, and 50 μM) or the respective vehicle (DMSO, 5 μM) for 6 days. (E) Schematic diagram for the comprehensive impact of STM2457 on METTL3 expression and activity in NSCLC. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
The effects of STM2457 may result from the inhibition of the catalytic activity and upregulation of protein expression on METTL3 (Fig. 4E). As an inhibitor of METTL3, STM2457 can “erase” the catalytic activity of METTL3 and increase its protein level through an unknown compensatory mechanism. At low concentrations or short time duration, there was prominent inhibition of METTL3 catalytic activity mediated by STM2457. Beyond this concentration or time node, the balance shifted toward STM2457-mediated upregulation of METTL3 protein expression. In general, regardless of the inactivation, the total expression of METTL3 was upregulated by STM2457, which could be regarded as a novel upregulator. Future studies should explore the application potential and optimize the structure of STM2457 based on its mechanism from the multi-omics analysis. The possibility of degrading METTL3 using a series of novel “degraders” de novo synthesized via peptide proteolysis-targeting chimeras, which prevent the METTL3 enzyme activity and downregulate its expression, should be also explored in the future [15].
3.2.2. Impact of STM2457 on transcriptomes in NSCLC
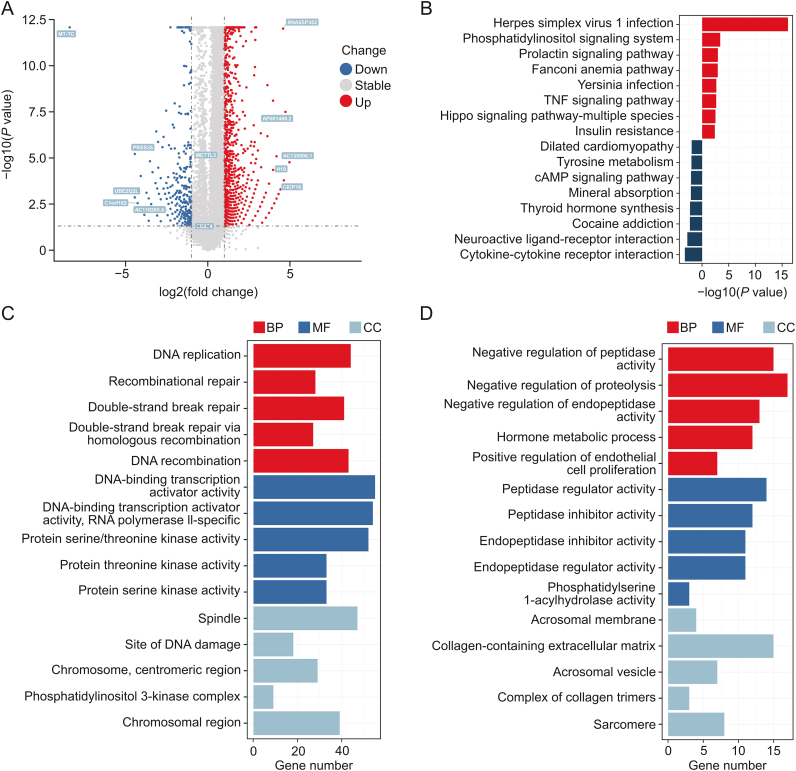
The m6A modification mediated by METTL3 is the most common mRNA methylation and plays an important role in RNA splicing, stability, and translation [16]. The effect of STM2457 on transcriptome and translatome in NSCLC has attracted significant interest. At the transcriptional level, the 1,713 upregulated genes (e.g., H19 and AC135506.1) and 507 downregulated genes (e.g., MT-TC and PRSS35) after STM2457 treatment were screened in A549 cells (Fig. 5A and Table S1). Notably, METTL3 (log2FC = 0.3775 < 1, P < 0.05), the target of STM2457, and PD-L1 (log2FC = −0.7204 > −1, P > 0.05), the ligand of the immune checkpoint receptor PD-1, were not significantly regulated. In the functional enrichment analysis (Table S2, Figs. 5B and C), the “tumor necrosis factor (TNF) signaling pathway” (gene ratio = 0.0362, P = 0.002) and “Hippo signaling pathway” (gene ratio = 0.0149, P = 0.003) were significantly enriched by upregulated genes in the KEGG analysis; both pathways have been explored by tumor research, including that related to NSCLC. The genes downregulated by STM2457 were significantly enriched in “cytokine-cytokine receptor interaction” (gene ratio = 0.093, P < 0.001), a large and complex pathway that involves the transforming growth factor-beta, TNF, and other cytokine families (Figs. 5B and D). Therefore, we believe that STM2457 may have significant effects on the NSCLC transcriptome through METTL3-mediated regulation of RNA splicing and stability, which may depend on the m6A methyltransferase activity of METTL3.
Fig. 5.
Impact of STM2457 on transcriptome of non-small cell lung cancer (NSCLC). (A) The volcano plot of differential expression analysis of RNA sequencing (RNA-seq) data acquired from A549 cells treated with STM2457 (5 μM) and the respective vehicle (dimethylsulfoxide (DMSO), 5 μM) for 3 days. (B) The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of the differentially expressed genes was analyzed using R/clusterProfiler packages. The Gene Ontology (GO) enrichment of (C) upregulated or (D) downregulated differentially expressed genes acquired from RNA-seq data of A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days was analyzed using the R/clusterProfiler package. TNF: tumor necrosis factor; cAMP: cyclic adenosine monophosphate; BP: biological processes; MF: molecular functions; CC: cell components.
3.2.3. Impact of STM2457 on translatome in NSCLC
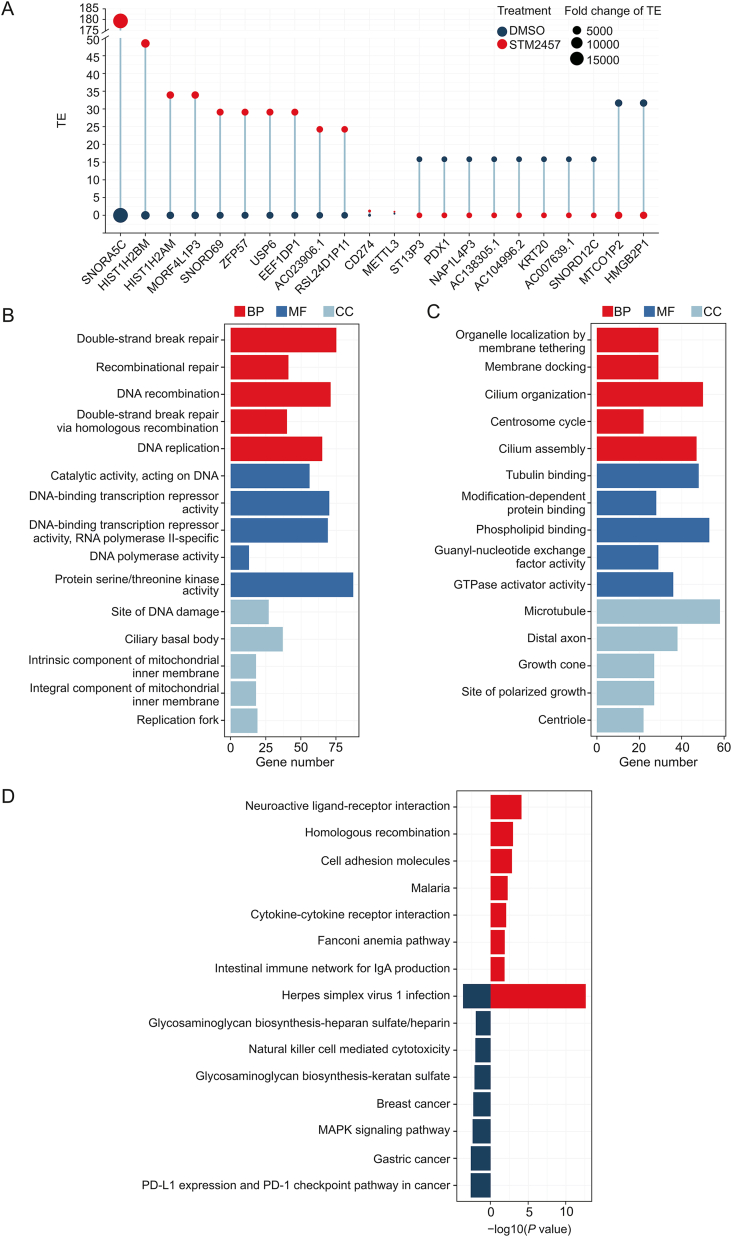
METTL3 interacts with the translation initiation machinery to promote translation independent of its methyltransferase activity [17]. Based on the comprehensive effects of STM2457 on METTL3, the effects of STM2457 on the translatome may be more important in NSCLC. After treatment with STM2457, 5,208 genes with a greater than two-FC of TE were screened based on the combined analysis of ribosome profiling (Ribo-seq) and RNA-seq in A549 cells (Table S3). Among the 3,371 upregulated genes, the TE of SNORA5C was most significantly increased (Fig. 6A). The TE of PD-L1 increased with DMSO exposure from 0.01 to 1.21 after STM2457 treatment (Fig. S3A), which is direct evidence for the regulatory effects of STM2457 on PD-L1 translation. Additionally, the TE of METTL3 was also increased by 2.1957-fold (Fig. S3B), suggesting a potential mechanism for METTL3 overexpression induced by STM2457. GO enrichment analysis for the aforementioned genes revealed their associations with DNA damage repair (Fig. 6B). However, 1,837 downregulated genes were mainly enriched in “organelle location by member tethering,” “tubulin binding,” and “microtubule” in terms of biological processes, molecular functions, and cell components, respectively (Fig. 6C), consistent with the inhibitory effects of STM2457 on the migration and invasion abilities of NSCLC. Except for PD-L1 with upregulated TE, it is worth mentioning that other genes including EGF and AKT1 involved in the “PD-L1 expression and PD-1 checkpoint pathway in cancer” pathway of the KEGG enrichment analysis were downregulated after STM2457 treatment (Fig. 6D and Table S4). Therefore, the regulatory effects of STM2457 on PD-L1 need to be explored further.
Fig. 6.
Impact of STM2457 on translatome of non-small cell lung cancer (NSCLC). (A) The dumbbell plot of the changes in translation efficiency (TE) of differentially translated genes (DTGs) with the top 10 upregulated (10 genes on the left) or downregulated TEs (10 genes on the right), as well as methyltransferase-like 3 (METTL3) and programmed death-ligand 1 (PD-L1), acquired from ribosome profiling (Ribo-seq) and RNA sequencing (RNA-seq) data of A549 cells treated with STM2457 (5 μM) and the respective vehicle (dimethylsulfoxide (DMSO), 5 μM) for 3 days. The Gene Ontology (GO) enrichment of DTGs with (B) upregulated or (C) downregulated TE was analyzed using the R/clusterProfiler package. (D) The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially translated genes (DTGs) acquired from Ribo-seq and RNA-seq data of A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days. BP: biological processes; MF: molecular functions; CC: cell components; GTPase: guanosine triphosphate hydrolase; MAPK: mitogen-activated protein kinase.
The polysome profiling data of METTL3-depleted or control HeLa cells [6] provided a new perspective to further understand the impact of STM2457 on the translation mechanisms through METTL3. The TE of almost all differentially translated genes (DTGs) was downregulated (Table S5), suggesting that the main effect of METTL3 deletion on the translation regulation is to induce its inhibition. Among 4,286 downregulated DTGs, the top three greatest TE FCs were for ROPN1L-AS1, PLEKHG6, and HHAT; the TE of RPPH1 most significantly increased as a member of 97 upregulated DTGs (Fig. 7A). Subsequently, the translatome data for STM2457 treatment and METTL3 depletion were combined and analyzed (Table S5). A subset of 659 DTGs, including PD-L1 and METTL3, have attracted significant interest due to their positive correlations with METTL3 expression (Fig. 7B). Consistent with STM2457 treatment, METTL3 depletion induced a decrease in PD-L1 TE from 3.58 to 1.19 and a decrease in METTL3 by 5.74-fold. Therefore, we speculate that the translation regulation of this gene set may be more dependent on METTL3 expression than its catalytic activity. Finally, the KEGG (Fig. 7C and Table S6) and GO (Figs. 7D and E) enrichment analyses found that the downregulated DTGs induced by METTL3 depletion were associated with fatty acid metabolism and cell cycle, respectively. Future research should focus on the specific underlying mechanism.
Fig. 7.
Impact of methyltransferase-like 3 (METTL3) depletion on translatome based on bioinformatics analysis of polysome profiling data acquired from HeLa cells. (A) The dumbbell plot of the changes in translation efficiency (TE) of differentially translated genes (DTGs) with the top 10 upregulated (10 genes on the left) or downregulated TEs (10 genes on the right), as well as METTL3 and programmed death-ligand 1 (PD-L1), acquired from polysome profiling data of METTL3-depleted and control HeLa cells. (B) Venn diagram of DTGs acquired from ribosome profiling (Ribo-seq) data of A549 cells treated with STM2457 and polysome profiling data of METTL3-depleted HeLa cells. (C) The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment of DTGs acquired from polysome profiling data of METTL3-depleted and control HeLa cells was analyzed using R/clusterProfiler package. The Gene Ontology (GO) enrichment of DTGs with (D) upregulated or (E) downregulated TE acquired from polysome profiling data of METTL3-depleted and control HeLa cells was analyzed using the R/clusterProfiler package. KD: knock down; NC: negative control; BP: biological processes; MF: molecular functions; CC: cell components; MHC: major histocompatibility complex; NAD: nicotinamide adenine dinucleotide; NADP: nicotinamide adenine dinucleotide phosphate.
3.2.4. Impact of STM2457 on m6A methylation modification in NSCLC
Although the current results indirectly indicate that STM2457 inhibits METTL3-mediated m6A methylation modification in NSCLC, the effects should be investigated in detail based on the meRIP-seq data. As the high-frequency m6A motif RGACH (R = G/A; H = A/C/U) (Fig. S4A), m6A peaks under the treatment of DMSO and STM2457 were significantly enriched on the motif “AGACU” (DMSO: P = 1 × 10−137 vs. STM2457: P = 1 × 10−66). Furthermore, with both treatments, m6A peaks were abundant in exons (34.84% vs. 35.28%), transcription termination sites (24.38% vs. 26.75%), and promoter-transcription start sites (21.53% vs. 23.43%), without significant between-group differences (Figs. S4B and C). Consistently, the effect of STM2457 on m6A peak enrichment was mainly downregulated (upregulated peaks: 67 vs. downregulated peaks: 258), suggesting an inhibitory effect of STM2457 on m6A methylation modification (Fig. 8A and Table S7). However, compared to METTL3 depletion (upregulated peaks: 2,602 vs. downregulated peaks: 2,576), the effect of STM2457 as the molecular inhibitor of METTL3 was relatively insignificant (Fig. 8B and Table S8). At the same time, the targets of inhibition were different between STM2457 treatment and METTL3 knockout. Among the 194 downregulated differentially m6A-modified genes (DMGs) enriched by 258 downregulated peaks, there were only 32 DMGs, of which m6A methylation is also inhibited by METTL3 knockout (Fig. 8C). GO enrichment analysis revealed the difference of function between downregulated DMGs induced by STM2457 treatment (Fig. 8E and Table S9) and METTL3 knockout (Fig. 8F). Subsequent exploration of the mechanisms underlying the aforementioned differences will enhance our understanding of the pharmacological action of STM2457, and more precise quantification of m6A based on liquid chromatography-tandem mass spectrometry and multi-omics analysis integrating metabolomics should be carried out.
Fig. 8.
Impact of STM2457 on N6-methyladenosine (m6A) RNA methylation modification of non-small cell lung cancer (NSCLC). (A) Scatter plot of m6A enrichment in mRNAs acquired from methylated RNA immunoprecipitation sequencing (meRIP-seq) data of A549 cells treated with STM2457 (5 μM) and the respective vehicle (dimethylsulfoxide (DMSO), 5 μM) for 3 days. m6A-containing mRNAs with significantly increased and decreased peak enrichment are highlighted (|log2 fold change (FC)| > 1 and P < 0.05). (B) Scatter plot of m6A enrichment in mRNAs acquired from meRIP-seq data of A549 negative control of METTL3 knockout (KO_NC) and A549 knockout of methyltransferase-like 3 (KO_METTL3). The m6A-containing mRNAs with significantly increased and decreased peak enrichment are highlighted (|log2FC| > 1 and P < 0.05). (C) Venn plot of downregulated differentially m6A-modified genes (DMGs) acquired from meRIP-seq data of A549 cells with STM2457 treatment and METTL3 knockout. (D) Venn diagram of upregulated, downregulated, and stable differentially m6A-modified genes (DMGs) acquired from meRIP-seq data and upregulated DTGs acquired from ribosome profiling (Ribo-seq) and RNA sequencing (RNA-seq) data in A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days. (E) The Gene Ontology (GO) enrichment of DMGs with hypo-down peaks acquired from meRIP-seq data of A549 cells with STM2457 treatment. (F) The GO enrichment of DMGs with hypo-down peaks acquired from meRIP-seq data of A549 cells with METTL3 knockout. BP: biological processes; MF: molecular functions; CC: cell components; UDP: uridine diphosphate.
To our interest, neither STM2457 treatment nor METTL3 knockdown significantly affected the m6A methylation status of METTL3 (Fig. S4D) and PD-L1 (Fig. S4E), which was different from the results of Ribo-seq. Similar to METTL3 and PD-L1, the translational efficiency was upregulated in a majority (2,163/3,371) of DTGs, whereas m6A methylation was not affected by STM2457 treatment (Fig. 8D). The aforementioned evidence suggests that METTL3-mediated m6A methylation may not be necessary for translation machinery. Additionally, METTL3 expression was upregulated by STM2457, which may have a profound effect on the translation of a majority of targets, consistent with the published reports that METTL3 translational regulation is independent of its enzyme activity [17].
3.3. Regulatory effects of STM2457 on PD-L1 expression in NSCLC
3.3.1. Model for the regulatory effects of STM2457 on PD-L1 expression in NSCLC
Admittedly, there is still a long way to go to completely reveal the mechanism underlying the effects of STM2457 in NSCLC. However, we could preliminarily construct a model for the regulation of STM2457 on the translation of PD-L1 based on the current evidence. Regardless of deactivation, total METTL3 was upregulated by STM2457 and interacted with the translation initiation machinery to circularize more mRNA. Then, more ribosomes were recruited for the circularized mRNA of PD-L1 to improve the TE of PD-L1, independent of its methyltransferase activity.
3.3.2. Validation of the effects of STM2457 on PD-L1 expression in NSCLC
PD-L1 on the NSCLC cellular surface interacts with PD-1 on T cells to inhibit activation, expansion, and effector functions of antigen-specific CD8+ T cells; the binding helps cancer cells to evade immune destruction. According to the extrinsic and intrinsic causes of PD-L1 expression in cancer cells, PD-L1 is classified into inducible and constitutive types, respectively. Among the extrinsic factors, IFN-γ is generally considered as the most prominent inducer of PD-L1 [18]. Therefore, we emphasized the differences in PD-L1 expression in location and cause during the validation of the effects of STM2457 on PD-L1 expression. Both constitutive and inducible PD-L1 on A549 (Fig. 9A) and H1975 (Fig. 9B) cells were significantly upregulated by STM2457 (0–5 μM) in a concentration-dependent manner, even though IFN-γ increases the baseline PD-L1 expression. Compared with PD-L1 expression on A549 cell surface without (Fig. 9C) or with induction of IFN-γ (Fig. 9D), the upregulation of constitutive (Fig. 9E) and inducible (Fig. 9F) PD-L1 expression by STM2457 on H1975 cell surface showed a more concentration-dependent effect.
Fig. 9.
Regulation of STM2457 on programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer (NSCLC). The regulation of STM2457 on the total expression of PD-L1 protein was detected using Western blot in (A) A549 and (B) H1975 cells treated with STM2457 (0, 0.05, 0.5, 5, and 50 μM) or the respective vehicle (dimethylsulfoxide (DMSO), 5 μM) for 3 days, with or without interferon-gamma (IFN-γ) induction (10 ng/mL) for 24 h. The regulation of STM2457 on the membrane expression of PD-L1 protein was detected using flow cytometry assay in A549 (C) without or (D) with IFN-γ induction (10 ng/mL) for 24 h and H1975 (E) without or (F) with IFN-γ induction (10 ng/mL) for 24 h. (G) The regulation of methyltransferase-like 3 (METTL3) knockout on the total expression of PD-L1 protein was detected using Western blot in wild type (WT), METTL3 knockout (KO), and the corresponding negative control (NC) A549 cells without or with IFN-γ treatment (10 ng/mL) for 24 h. The regulation of METTL3 knockout on the membrane expression of PD-L1 protein was detected using flow cytometry assay in METTL3 KO (KO_METTL3) and the corresponding NC (KO_NC) A549 cells (H) without or (I) with IFN-γ treatment (10 ng/mL) for 24 h. (J) The regulation of METTL3 with defective catalytic activity on PD-L1 expression was detected using Western blot in A549 cell lines with overexpression of WT (OE_METTL3), catalytic defect (OE_METTL3_Mut), and the corresponding NC (OE_NC and OE_Mut_NC). GAPDH: glyceraldehyde 3-phosphate dehydrogenase; FL4-A: fluorescent light channel 4-area; APC-A: allophycocyanin-area ; OE: overexpression; Mut: mutation.
To clarify the role of METTL3 methyltransferase activity in the abovementioned regulation, we knocked out METTL3, upregulated wild-type METTL3, or transfected METTL3 with catalytic defects in A549. METTL3 depletion inhibited total (Fig. 9G) and membrane expression of PD-L1 without (Fig. 9H) or with IFN-γ induction (Fig. 9I). Interestingly, knockout of METTL3 made the A549 cell line more insensitive to STM2457 (Fig. S5A), and STM2457 further reduced the relatively low global m6A modification at the mRNA level (Fig. S5B) even in A549 with METTL3 depletion, between which the potential mechanism needs to be further explored. Moreover, transfected wild-type METTL3 and METTL3 with catalytic defects increased the total PD-L1 expression, which supports that the methyltransferase activity of METTL3 was unnecessary in the aforementioned model (Fig. 9J). Finally, in vivo STM2457 could upregulate PD-L1 and increase METTL3 expression (Fig. 3B). PD-L1 upregulation converts cold tumors into hot tumors [19], and STM2457 improves efficacy with immunotherapy. Although several recent in vivo studies confirmed that STM2457 plus anti-PD-1 showed promising antitumor efficacy against cervical squamous cell carcinoma [20] and colorectal cancer [21], all of them were based on its role as a small-molecule inhibitor of METTL3 while ignoring its more powerful translation regulation of immune checkpoint PD-L1.
4. Conclusions
In conclusion, STM2457 is a potential novel suppressor based on its inhibitory effect on tumor progression, ability to overcome heterogeneity based on its impact on the translatome, and ability to improve the immunotherapy benefits based on PD-L1 upregulation in NSCLC. However, further studies are needed to confirm the in vivo effects of STM2457 and explore its multi-omics characteristics. More experimental evidence is needed to support the theoretical possibility that STM2457 can overcome tumor heterogeneity in NSCLC, including the patient-derived tumor xenograft model to simulate heterogeneous environments in vivo and single-cell sequencing to explore the effects of STM2457 on heterogeneity. Additionally, in vivo and in vitro studies should be conducted on the combination of PD-1/PD-L1 antibodies. The current results provide an excellent basis for targeting METTL3 as a therapeutic strategy for NSCLC.
CRediT author statement
Han Xiao: Conceptualization, Methodology, Software, Resources, Investigation, Data curation, Formal analysis, Visualization, Writing - Original draft preparation, Project administration; Rong Zhao: Investigation, Validation, Data curation, Visualization, Writing - Reviewing and Editing; Wangyang Meng: Investigation, Validation, Writing - Reviewing and Editing; Yongde Liao: Conceptualization, Resources, Writing - Reviewing and Editing, Project administration, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No.: 82072593). We would like to thank the research team of Dr. Eliza Yankova, who initially synthesized STM2457 and published its structure. We are also grateful to Dr. Junho Choe and his research team for their pioneering work in the field of METTL3-mediated translation regulation.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.04.009.
Contributor Information
Han Xiao, Email: 13260536972@163.com.
Yongde Liao, Email: liaotjxw@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
The effect of STM2457 on non-small cell lung cancer (NSCLC) migration. The effect of STM2457 on NSCLC migration was evaluated using wound-healing assay for 24 h in A549 (A) and H1975 (B) cells pretreated with STM2457 (5 μM) for 1, 3, and 6 days or the respective vehicle DMSO for 6 days.
The effect of STM2457 on non-small cell lung cancer (NSCLC) apoptosis. The regulation of STM2457 on apoptosis-associated proteins was detected using western blot in A549 (A) and H1975 (B) treated with STM2457 (1 and 5 μM) or the respective vehicle DMSO for 3 days. The analysis and comparison for the relative protein expression of cleaved PARP (C and D) and Caspase-3 (E and F) and the ratio of Bcl-2 to Bax (G and H) were performed based on gray values in A549 and H1975, respectively.
The mapping to PD-L1 (C) or METTL3 (D) of reads acquired from Ribo-seq and RNA-seq data in A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days was performed using the Integrative Genomics Viewer (IGV) tool.
Analysis of meRIP-seq data from A549 cells treated with STM2457. (A) Motif analysis of m6A modification peaks acquired from meRIP-seq data of A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days. (B) In five transcript segments, the fraction of m6A modification peaks acquired from meRIP-seq data of A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days. (C) Distribution profile of m6A modification peak reads across all mRNAs acquired from meRIP-seq data of A549 cells with STM2457 treatment and METTL3 knockout. The mapping to METTL3 (D) or PD-L1 (E) of reads acquired from meRIP-seq data of A549 cells with STM2457 treatment and METTL3 knockout was performed using the Integrative Genomics Viewer (IGV) tool.
The effects of STM2457 in A549 cells with METTL3 knockout. (A) Dose–response curves of STM2457 treatment for 6 days in METTL3 KO (KO_METTL3), the corresponding NC (KO_NC) A549 cells, and wild-type (WT) A549 cells. (B) The effect of STM2457 on the global m6A RNA modification levels was detected using dot blot assay in A549 KO_METTL3 and KO_NC treated with STM2457 (5 μM) or the respective vehicle DMSO for 3 days.
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z., Fillmore C.M., Hammerman P.S., et al. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 4.Reck M., Remon J., Hellmann M.D. First-line immunotherapy for non-small-cell lung cancer. J. Clin. Oncol. 2022;40:586–597. doi: 10.1200/JCO.21.01497. [DOI] [PubMed] [Google Scholar]
- 5.Bhat M., Robichaud N., Hulea L., et al. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 6.Choe J., Lin S., Zhang W., et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng C., Huang W., Li Y., et al. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020;13:117. doi: 10.1186/s13045-020-00951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni Z., Sun P., Zheng J., et al. JNK signaling promotes bladder cancer immune escape by regulating METTL3-mediated m6A modification of PD-L1 mRNA. Cancer Res. 2022;82:1789–1802. doi: 10.1158/0008-5472.CAN-21-1323. [DOI] [PubMed] [Google Scholar]
- 9.Yankova E., Blackaby W., Albertella M., et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593:597–601. doi: 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Q.-C., Tien Y.C., Shi Y.-H., et al. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m6A-YTHDF2-dependent manner. Oncogene. 2022;41:1622–1633. doi: 10.1038/s41388-022-02185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Z.-W. Zhang, X. Teng, F. Zhao, et al., METTL3 regulates m6A methylation of PTCH1 and GLI2 in Sonic hedgehog signaling to promote tumor progression in SHH-medulloblastoma, Cell Rep. 41 (2022), 111530. [DOI] [PubMed]
- 12.Loayza-Puch F., Rooijers K., Buil L.C., et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016;530:490–494. doi: 10.1038/nature16982. [DOI] [PubMed] [Google Scholar]
- 13.Leiser D., Pochon B., Blank-Liss W., et al. Targeting of the MET receptor tyrosine kinase by small molecule inhibitors leads to MET accumulation by impairing the receptor downregulation. FEBS Lett. 2014;588:653–658. doi: 10.1016/j.febslet.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 14.Bates S.E. Epigenetic therapies for cancer. N. Engl. J. Med. 2020;383:650–663. doi: 10.1056/NEJMra1805035. [DOI] [PubMed] [Google Scholar]
- 15.Chamberlain P.P., Hamann L.G. Development of targeted protein degradation therapeutics. Nat. Chem. Biol. 2019;15:937–944. doi: 10.1038/s41589-019-0362-y. [DOI] [PubMed] [Google Scholar]
- 16.Lan Q., Liu P.Y., Haase J., et al. The critical role of RNA m6A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 17.Lin S., Choe J., Du P., et al. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C., Mezzadra R., Schumacher T.N. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu M., Huang Q., Xie Y., et al. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J. Hematol. Oncol. 2022;15:24. doi: 10.1186/s13045-022-01242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu R., Wei Y., He C., et al. Integrative analyses of m6A regulators identify that METTL3 is associated with HPV status and immunosuppressive microenvironment in HPV-related cancers. Int. J. Biol. Sci. 2022;18:3874–3887. doi: 10.7150/ijbs.70674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H., Pan Y., Zhou Q., et al. METTL3 inhibits antitumor immunity by targeting m6A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology. 2022;163:891–907. doi: 10.1053/j.gastro.2022.06.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The effect of STM2457 on non-small cell lung cancer (NSCLC) migration. The effect of STM2457 on NSCLC migration was evaluated using wound-healing assay for 24 h in A549 (A) and H1975 (B) cells pretreated with STM2457 (5 μM) for 1, 3, and 6 days or the respective vehicle DMSO for 6 days.
The effect of STM2457 on non-small cell lung cancer (NSCLC) apoptosis. The regulation of STM2457 on apoptosis-associated proteins was detected using western blot in A549 (A) and H1975 (B) treated with STM2457 (1 and 5 μM) or the respective vehicle DMSO for 3 days. The analysis and comparison for the relative protein expression of cleaved PARP (C and D) and Caspase-3 (E and F) and the ratio of Bcl-2 to Bax (G and H) were performed based on gray values in A549 and H1975, respectively.
The mapping to PD-L1 (C) or METTL3 (D) of reads acquired from Ribo-seq and RNA-seq data in A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days was performed using the Integrative Genomics Viewer (IGV) tool.
Analysis of meRIP-seq data from A549 cells treated with STM2457. (A) Motif analysis of m6A modification peaks acquired from meRIP-seq data of A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days. (B) In five transcript segments, the fraction of m6A modification peaks acquired from meRIP-seq data of A549 cells treated with STM2457 (5 μM) and the respective vehicle (DMSO, 5 μM) for 3 days. (C) Distribution profile of m6A modification peak reads across all mRNAs acquired from meRIP-seq data of A549 cells with STM2457 treatment and METTL3 knockout. The mapping to METTL3 (D) or PD-L1 (E) of reads acquired from meRIP-seq data of A549 cells with STM2457 treatment and METTL3 knockout was performed using the Integrative Genomics Viewer (IGV) tool.
The effects of STM2457 in A549 cells with METTL3 knockout. (A) Dose–response curves of STM2457 treatment for 6 days in METTL3 KO (KO_METTL3), the corresponding NC (KO_NC) A549 cells, and wild-type (WT) A549 cells. (B) The effect of STM2457 on the global m6A RNA modification levels was detected using dot blot assay in A549 KO_METTL3 and KO_NC treated with STM2457 (5 μM) or the respective vehicle DMSO for 3 days.