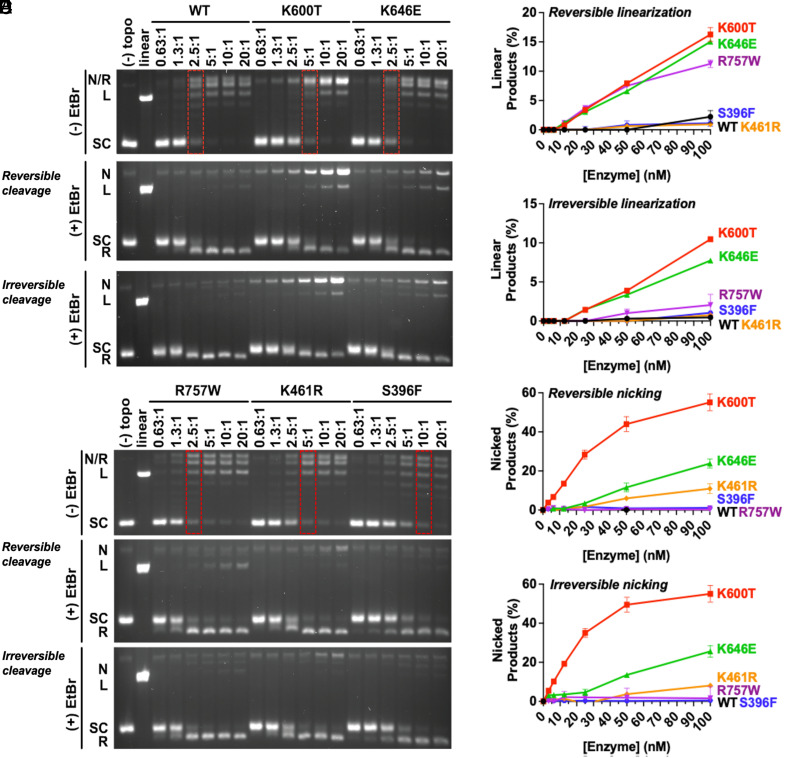
Fig. 2.
EtopHS hTOP2β variants display elevated nicking and linearization activities in vitro in the absence of drug. (A and B) Representative agarose gels of supercoil relaxation and cleavage reactions conducted in the absence of etoposide for purified mutant proteins identified from the EtopHS screen. Enzyme titrations are denoted as the molar ratio of hTOP2β dimer to plasmid DNA. Red dotted boxes indicate enzyme concentrations necessary to achieve levels of supercoil relaxation comparable to wild type (WT). The Middle panels of (A) and (B) were quenched to reveal reversible cleavage, whereas the bottom panels were quenched to reveal irreversible cleavage. (C–F) Nicking and linearization of DNA during the relaxation of negatively supercoiled plasmid by EtopHS hTOP2β variants are compared to wild-type hTop2β. Reactions were stopped by (C and E) 1% SDS alone to trap reversible cleavage complexes, or by (D and F) 1% SDS and 50 mM EDTA to isolate irreversible cleavage complexes. Plots show a summary of DNA linearization and nicking activity by EtopHS hTOP2β mutants, as quantified by densitometry of agarose-gel-resolved cleavage reactions. Reversible and irreversible cleavage complex formation is enhanced by several etoposide-hypersensitive hTOP2β mutations. Error bars represent the SD between three experimental replicates.