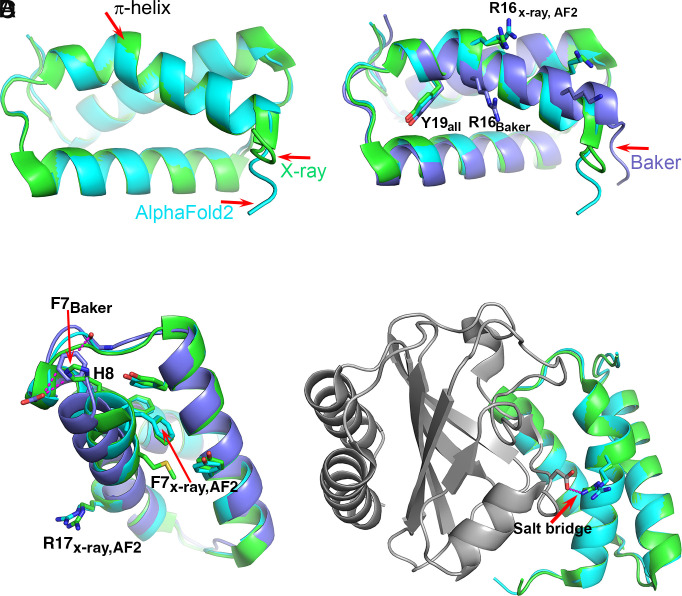
Fig. 2.
The T1046S1 (antiholin) π-helix. (A) Superposition of the crystal (green) and the AF2 (sky blue) structures. Both contain a short π-helix. (B) Superposition of the crystal (green), AF2 (sky blue), and Baker’s group (lavender) structures. The Baker group structure has a continuous α-helix with no π interruption, resulting in a different position of Arg16. (C) The environment of Phe7 in the three structures. In the crystal and AF2 structures, Phe7 is located within a hydrophobic core and His8 is exposed to solvent, whereas in the Baker’s group structure, Phe7 is located in approximately the same position as His8 in the crystal structure and interacts unfavorably with a backbone CO and a carboxyl group. The location of Arg16 on the experimental and AF2 structures is indicated. (D) Superposition of the crystal (green) and AF2 (sky blue) structures in the context of the complex with holin (gray). Arg16 forms a salt bridge with an aspartic acid on the partner holin protein. Without the π-helix segment, that key intermolecular interaction cannot be made. In this case, the AF2 structure includes a rare motif that is stabilized by interactions nine residues away, and that is critical to function.