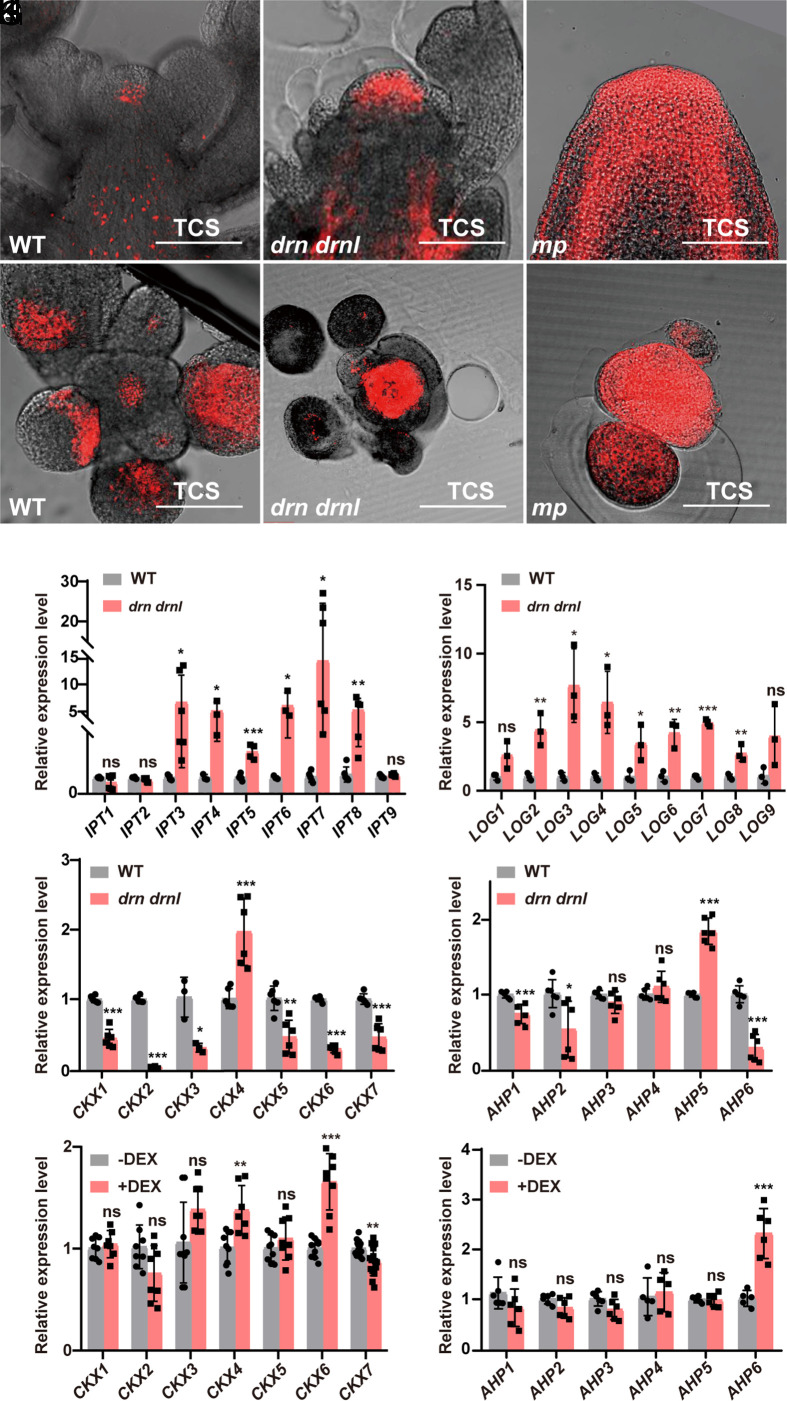
Fig. 2.
Cytokinin signaling is disrupted in drn drnl and mp mutants. (A–F) Cytokinin signaling detected by TCS::dTomato in the inflorescence apexes of Col-0 (A and D), drn drnl (B and E), and mp (C and F) plants with longitudinal (A–C) and transverse sections (D–F). n ≥ 20 shoot apexes per genotype were observed with similar results. (Scale bars, 100 μm.) (G–J) Expression levels of cytokinin-related genes in biosynthesis, degradation, and signal transduction pathways in the SAM of the drn drnl mutant, including IPTs (G), LOGs (H), CKXs (I) and AHPs (J). The data are shown as mean ± SD; n ≥ 3 biological replicates, two-tailed Student’s t tests, *P < 0.05, **P < 0.01, ***P < 0.001; ns, no significant difference. (K and L) Expression levels of CKXs (K) and AHPs (L) in the inflorescence apexes of UBQ10::DRNL-GR plants with or without DEX induction in the presence of cycloheximide using qRT–PCR. The data are shown as mean ± SD; n ≥ 5 biological replicates, two-tailed Student’s t tests, **P < 0.01, ***P < 0.001; ns, no significant difference.