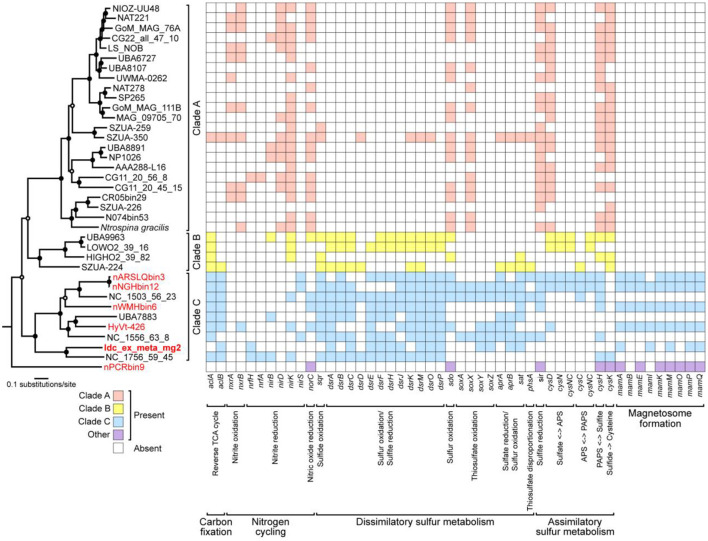
Figure 5.
A maximum-likelihood tree of 120 concatenated bacterial single-copy marker proteins from Nitrospinae genomes, as well as a heat map of Nitrospinae genomes showing the presence or absence of genes involved in carbon, nitrogen, and sulfur metabolisms as well as magnetosome genes. Genes involved in magnetosome formation were annotated by FeGenie. Other genes were annotated by METABOLIC and DiSCo. A concatenated sequence obtained in this study is indicated in bold letters, whereas concatenated sequences containing MGCs are indicated in red letters. At nodes in the tree, filled and open black circles represent 1,000 pseudoreplicate bootstrap values higher than 75 and 50%, respectively. The absence of aclAB in Clade A may be due to the low similarity of aclAB sequences in Clade A genomes to known aclAB sequences.