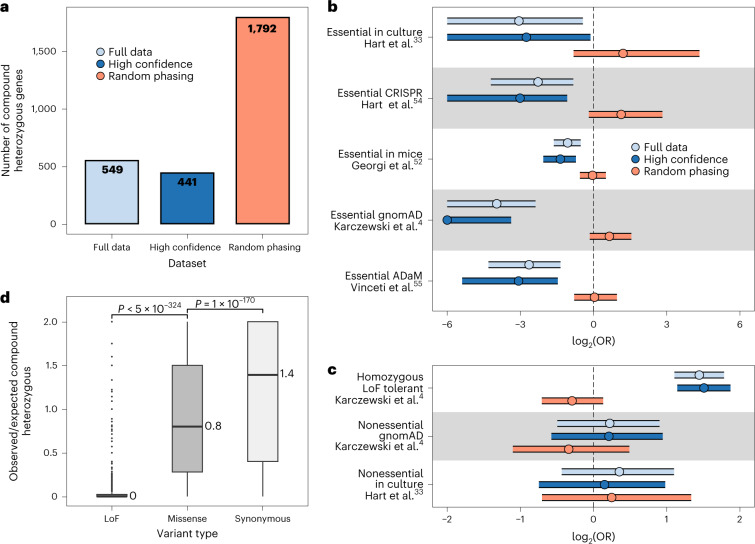
Fig. 3. Compound heterozygous identification in the UKB WES data phased with SHAPEIT5.
a, Number of genes with at least one individual with compound heterozygous LoF variants across several categories: Full data, all LoF variants in the study, except singletons; High confidence, LoF variants excluding calls with phasing confidence score <0.99; and Random phasing, shuffling phasing of all LoF variants (once). b, Two-way Fisher’s exact test odds ratios ± 95% confidence interval (log2-scaled) of compound heterozygous genes versus noncompound heterozygous genes presence in several lists of essential genes (Methods). Background is composed of 3,018 genes with ≥2 LoF mutations; x axis is capped at –6. c, Same as b but across lists of nonessential or LoF tolerant genes. d, Ratio between the number of individuals with compound heterozygous events and the expected number of individuals given the number of variants, per gene. Missense (n = 14,336 genes) and synonymous (n = 9,816 genes) events are shown in addition to LoF events (n = 2,150 genes) as a comparison. The length of the box corresponds to the interquartile range (IQR) with the center line and values corresponding to the median, and the upper and lower whiskers represent the largest or lowest value no further than 1.5× IQR from the third and first quartile, respectively. P values between categories correspond to two-sided Wilcoxon test P values.