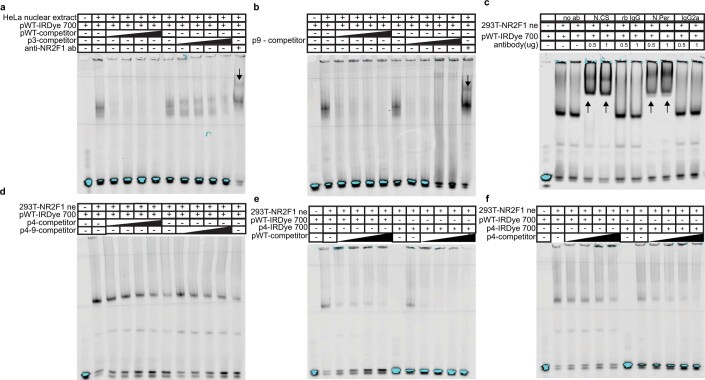
Extended Data Fig. 4. Additional EMSA data.
Electrophoretic mobility shift assay (EMSA) to confirm the interaction of NR2F1 with cRE2 sequence and to test whether HCFP1 SNVs attenuated this interaction in vitro. Blots are unmodified. Oligonucleotide probes containing the Cluster A and B region conjugated to a IRDye 700 fluorophore and competed with WT or mutant non-conjugated probes were designed. As per Fig. 4, EMSA results showing the effect of SNVs on NR2F1 binding (293T-NR2F1 ne denotes nuclear extract from NR2F1 transfected 293T cells) in the presence of increasing molar excess (25x-50x-100x-200x as denoted by black slope) of WT (pWT) or mutant (pMut) competitor probe compared to hot probe (pWT-IRDye 700). (a-b) EMSA for Cluster A p3 (a) and Cluster B p9 (b) using HeLa nuclear extract (refer to Fig. 4j for probe maps) (n = 4). The shifted band in the second lane of each gel is abolished with the addition of small amounts of WT or Cluster B p9 competitor. The Cluster A p3 is a less efficient competitor, suggesting that the variant alters the binding of a TF to the DNA. The addition of an anti-NR2F1 antibody causes a supershift of the TF-conjugated probe complex, indicating that this interaction is mediated by NR2F1. (c) A stronger shift is obtained with nuclear extract from NR2F1-transfected 293T cells. Specific supershift is observed using two different commercial NR2F1 antibody preparations (N.CS: Cell Signaling; N.Per: Perseus) each at two concentrations (0.5 ug and 1 ug). No supershift was observed using two isotype-specific controls (rabbit IgG for the N.CS antibody and IgG2a for the N.Per antibody), (n = 4). (d) No additional effects on competition are observed combining a variant in Cluster A and a variant in Cluster B on a single competitor probe (p4-p9 probe compared to p4 only (n = 6)). (e) Unlabeled pWT competes with labeled p4 more effectively than with labeled pWT (n = 2). (f) Unlabeled p4 does not compete well with labeled pWT. Comparing unlabeled pWT and unlabeled p4, the former competes better with labeled p4 (n = 2).