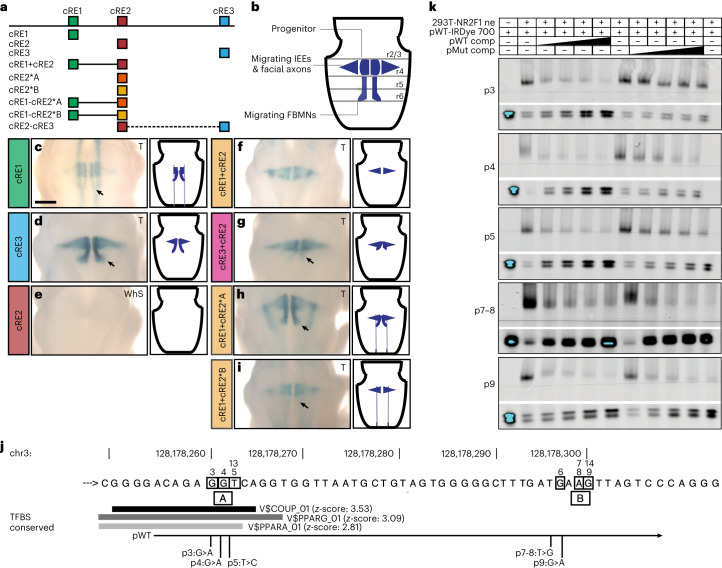
Fig. 4. Cluster A SNVs impair cRE2-mediated silencing in a reporter expression assay in vivo and reduce NR2F1 binding in vitro.
a,b, Schematics for in vivo lacZ reporter assay constructs (a) and hindbrain β-galactosidase expression viewed dorsally through the fourth ventricle (b). In b, midline ovals denote IEE/FBMN progenitors, triangles denote migrating IEEs and leg-like columns denote migrating FBMNs that are highlighted by black arrows in c, d and g–i. c–i, Selected images of ectopic β-galactosidase in transfected embryos (left) and schema (right): cRE1 alone (c, n = 13), cRE3 alone (d, n = 6) cRE2 alone (e, n = 8), cRE1 with cRE2 (f, n = 10), cRE3 with cRE2 (g, n = 7), cRE1 with cRE2 carrying Cluster A variants (h, n = 13) and cRE1 with cRE2 carrying Cluster B variants (i, n = 8). The asterisk denotes a mutant cluster. Scale bar (c), 500 μm and applies to c–i. Additional images are shown in Extended Data Fig. 3. j, Partial cRE2 sequence, as per Fig. 1. Gray horizontal bars denote overlap with in silico, conserved, transcription-binding consensus sequences from TRANSFAC (indicated by $). The shade of gray correlates with a prediction z-score. WT (pWT) and mutant (pMut) EMSA probes are aligned below. TFBS, TF-binding sites. k, EMSA results showing the effect of SNVs on NR2F1-binding activity from transfected nuclear extract (293T-NR2F1 ne) in the presence of increasing molar excess (25× to 50× to 100× to 200× as denoted by black slope) of pWT or pMut competitor ‘cold’ probes compared with conjugated ‘hot’ probe (pWT-IRDye 700). For each SNV: NR2F1 binding (upper gel); free probe (bottom gel, lower and upper bands reflect unannealed and annealed probe, respectively). In all five experiments, pWT shows decreasing NR2F1 binding and increasing free probes. Cluster A variant competitor probes (p3, p4 and p5) compete less well than pWT for NR2F1 binding (more NR2F1 shifted and less free probe available). Cluster B variants (p7–8 and p9), where no NR2F1 binding is expected, show no substantial effect. The same trend was observed in replicate experiments: WT = 11; p3 = 5; p4 = 8; p5 = 4; p7-8 = 3; and p9 = 7. Full gels are given in Source data.