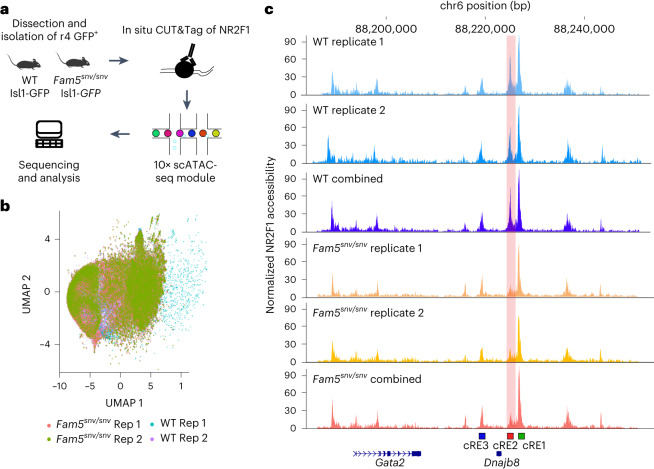
Fig. 5. NR2F1 binds cRE2 in E10.5 r4MNs and binding is reduced by Fam5 SNV.
a, Schematic representation of single-cell CUT&Tag of E10.5 WT and Fam5snv r4 Isl1+ neurons targeting NR2F1. scATAC-seq, single-cell assay for transposase-accessible chromatin with high-throughput sequencing. b, UMAP embedding of NR2F1 single-cell CUT&Tag experiment for two Fam5SNV/SNV (replicate (Rep) 1 = 2,274 cells, Rep 2 = 2,740 cells) and two WT (Rep 1 = 2,572 cells, Rep 2 = 1,377) age-matched biological replicates. c, Pseudobulk single-cell CUT&Tag profile of NR2F1 around the Gata2 regulatory region shown as individual and combined replicates (WT in blue and Fam5SNV/SNV in red–yellow). Location of the cREs relative to mouse Gata2 and Dnajb8 is shown. Note the reduction in the height of the cRE2 peaks (vertical pink shading) in the Fam5SNV/SNV replicates.