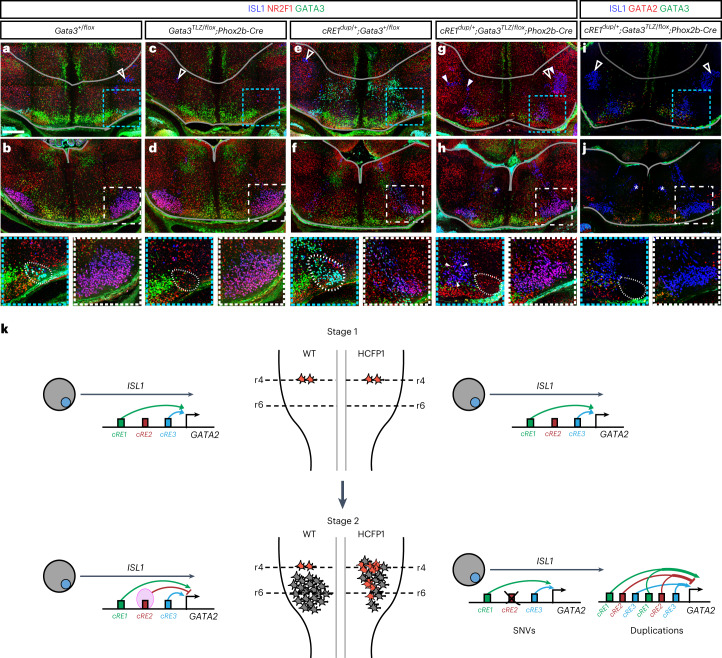
Fig. 8. Combining cRE1dup with Gata3 conditional inactivation partially rescues the HCFP1 phenotypes.
a–h, ISL1 (blue), NR2F1 (red) and GATA3 (green) immunofluorescent staining of hindbrain cross-sections at r4 (top row) and r6 (middle row) axial levels in E14.5 Gata3flox/+;Phox2b-Cre− WT (a,b), Gata3tlz/flox;Phox2b-Cre+ conditional Gata3 knockout (c,d), cRE1dup/+;Gata3flox/+ duplication (e,f) and cRE1dup/+;Gata3tlz/flox;Phox2b-Cre+ rescue (g,h) embryos. i,j, A rescue embryo with ISL1 (blue), GATA2 (red) and GATA3 (green) immunofluorescence (for WT and cRE1dup/+ comparators, see Fig. 7a,c,e,g). Dotted blue squares surround IEE OCNs in a, c, e, g and i and are magnified (bottom row). Dotted white squares marking the right facial nucleus are boxed in b, d, f, h and j and magnified (bottom row). Rescue embryos lack OCNs (g,i) and form an FBMN nucleus (h,j) intermediate in cross-sectional area between WT (b) and cRE1dup/+ (f) embryos. White arrows in magnification of g highlight r4 ISL1ON;NR2F1ON FBMNs. White open arrowheads show trigeminal motor neurons and asterisks the abducens nucleus. Dorsal and ventral borders of the hindbrain are outlined in gray. Scale bar, 200 μm in a applies to a–j (n = 3 (a,b), 3 (c,d), 4 (e,f), 3 (g,h) and 5 (i,j) embryos). k, Model depicting the effect of HCFP1 variants. Stage 1: in both WT (left side) and HCFP1 (right side) hindbrains, early born r4MN progenitors express Gata2, driven in part by cRE1 and cRE3 enhancers, and assume an IEE identity (red cells). Stage 2: in WT, NR2F1 (pink oval) binds to cRE2 in later-born r4MNs, silencing GATA2 and directing these cells to an FBMN identity (gray cells). In HCFP1, cRE2 SNVs disrupt NR2F1 binding (demarcated with X) and unimpeded cRE1 and cRE3 enhancers drive GATA2 expression in later-born r4MNs. Duplications of cRE1, cRE2 and cRE3 generate a net increase in GATA2 enhancer level, similarly expanding GATA2 expression. Either will increase IEEs at the expense of FBMNs, deplete the FBMN progenitor pool and result in CFP.