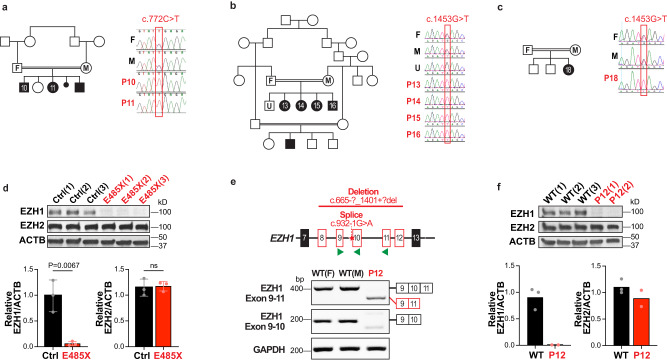
Fig. 2. Biallelic EZH1 variants cause loss of function.
a–c Pedigree and Sanger sequencing showing segregation of variants with the diseases in consanguineous families with (a), 2 affected children (p10-11) harboring the homozygous nonsense EZH1 c.772C>T; p.R258X variant (note that the youngest brother in the family was not included in the study) (b), 4 affected children (P13-16) harboring the homozygous nonsense EZH1 c.1453G>T: p.E485X variant. and (c), a child (P18) from an unrelated family with shared haplotype and EZH1 c.1453G>T: p.E485X variant. “F” indicates father, “M” mother and “U” unaffected family members. d Western blot analysis of EZH1, EZH2 and ACTB showing significant loss of EZH1 in hPSCs harboring the homozygous EZH1 p.E485X variant compared to isogenic controls. Graph shows mean ± SD of relative levels quantified by band densitometry in n = 3 independent clones. Two-sided unpaired t test. e Schematic representation of EZH1 exon 7-13, indicating the location of deletion and splice variants in P12 and the primers used for RT-PCR with green arrowheads (top). RT-PCR results showing undetectable EZH1 exon 10 containing transcripts in P12 lymphoblastoid cells compared to two unrelated wild type lymphoblastoid cells (WT(F) = females; WT(M) = male) and GAPDH as loading control (bottom). f Western blot analysis showing undetectable EZH1 levels and intact EZH2 in P12 cells compared to unrelated wild type (WT) cells. ACTB is shown as loading control. Graphs show mean relative EZH1/ACTB and EZH2/ACTB levels quantified by band densitometry in n = 3 WT cell lines and n = 2 P12 independent clonal cell lines. Statistical comparisons are not shown due to small sample size. Source data are provided as a Source Data file.