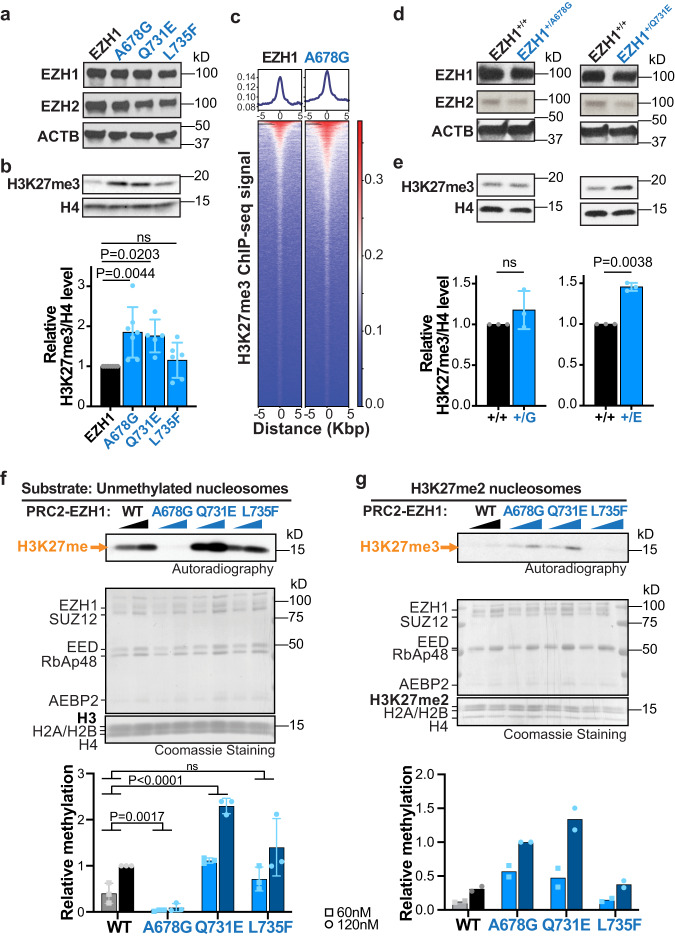
Fig. 3. Heterozygous missense variants cause EZH1 gain of function leading to hypermethylation of H3K27.
a, b Western blot analysis of EZH1, EZH2 (a) and H3K27me3 (b) in ReNcells transiently expressing either wild type or indicated EZH1 missense variants. ACTB or H4 are shown as loading controls. Graph shows mean ± SD of H3K27me3/H4 levels quantified by band densitometry in n = 7 EZH1, n = 7 p.A678G, n = 5 p.Q731E and n = 6 p.L735F independent transductions. ns = non-significant. One-way ANOVA with Dunnett’s post hoc analysis test for multiple comparisons. c Enrichment plots showing average signal of H3K27me3 in ChIPseq peaks (top) and heatmaps showing normalized H3K27me3 ChIPseq intensities (bottom) ±5 kb around the center of the peak in EZH1 or A678G expressing ReNcells. Plots represent data combined from 3 independent transductions. Two-sided paired t-test of signal in H3K27me3 peaks indicates statistically significant increase of H3K27me3 in A678G (p-value < 2.2e-16). d, e Western blot analysis of EZH1, EZH2 (d), and H3K27me3 (e) in 4-week old neurons derived from hPSCs carrying EZH1 p.A678G or p.Q731E variants in heterozygosity (EZH1+/A678G (+/G), EZH1+/Q731E, (+/E)) or their isogenic controls (EZH1+/+). ACTB or H4 are shown as loading controls. Graph shows mean ± SD of relative H3K27me3/H4 levels quantified by band densitometry in n = 3 independent differentiations. ns = non-significant. Two-sided paired t test. f, g Autoradiography and Coomassie stains of HMT assay reactions using two increasing concentrations of PRC2 complexes and unmethylated nucleosomes (f) or nucleosomes with dimethylated H3K27 (H3K27me2) as substrate (g). Graphs show mean ± SD of relative methylation levels quantified by band densitometry in n = 3 (f) or n = 2 (g) independent assays. ns = non-significant. Two-way ANOVA with Dunnett’s post hoc analysis test for multiple comparisons for main variant effect (f). Statistical comparisons are not shown for graph (g) due to small sample size. Source data are provided as a Source Data file.