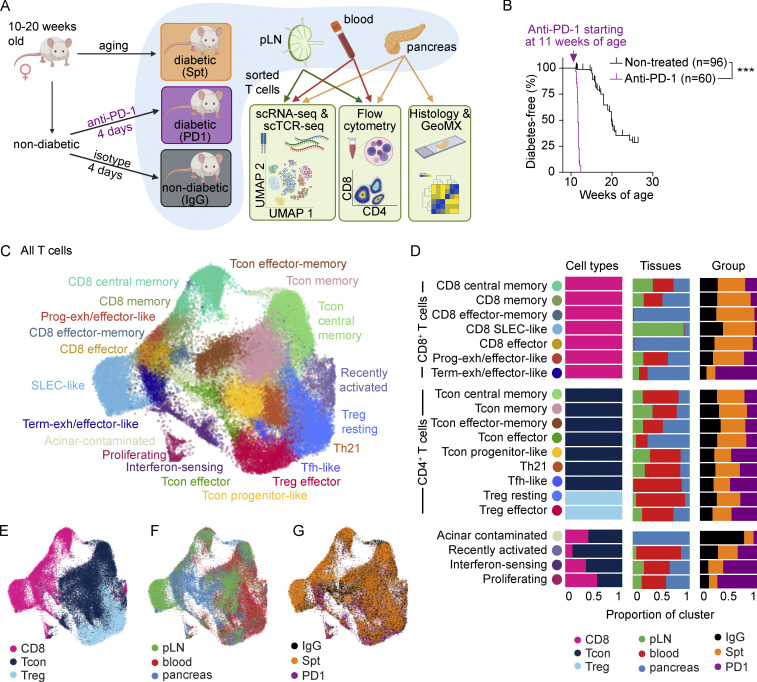
Figure 1.
Comparison of single-cell transcriptional landscape of CD4+ and CD8+ T cells during spontaneous and anti-PD-1-induced T1D in NOD mice. (A) Study design for multisite analysis of IgG, Spt, or PD1 mice. (B) Percent diabetes-free NOD mice following administration of two doses of anti-PD-1 antibody (purple) or isotype control antibody (black) every other day beginning at 11 wk of age. Kaplan–Meier survival curves and results from a Mantel-Cox log-rank test (P < 1 × 10−15) are shown. (C) Clustering and UMAP visualization of integrated CD4+ and CD8+ T cell data from paired blood, pLN, and pancreas from three mice per treatment group (IgG, PD1, and Spt). Colors denote transcriptional clusters, labeled with functional annotations. See Materials and methods for more details on functional annotations, and Table S1 for full list of upregulated genes per cluster. (D) Stacked bar plots showing the proportion of cell types (Tcon, Treg, and CD8+ T cells), tissues, and treatment groups within each cluster. Definitions used to classify cells as CD8+, Tcon, or Treg are indicated in the Materials and methods. The term “Tcon” specifically refers to CD4+ Foxp3− T cells. Most clusters were exclusively one cell type (e.g., CD8+ T cells, Tcons, or Tregs). However, some clusters (e.g., bottom four rows) contained a mix of CD8+ T cells and Tcons. (E–G) UMAP visualization of the distribution of all T cells based on (E) cell type classification, (F) tissues, and (G) treatment groups. Asterisks indicating significance: ***, P < 0.001.