Abstract
β-Catenin (Ctnnb1) has been shown to play critical roles in the development and maintenance of epithelial cells, including the retinal pigment epithelium (RPE). Ctnnb1 is not only a component of intercellular junctions in the epithelium, it also functions as a transcriptional regulator in the Wnt signaling pathway. To identify which of its functional modalities is critically involved in mouse RPE development and maintenance, we varied Ctnnb1 gene content and activity in mouse RPE lineage cells and tested their impacts on mouse eye development. We found that a Ctnnb1 double mutant (Ctnnb1dm), which exhibits impaired transcriptional activity, could not replace Ctnnb1 in the RPE, whereas Ctnnb1Y654E, which has reduced affinity for the junctions, could do so. Expression of the constitutively active Ctnnb1∆ex3 mutant also suppressed the development of RPE, instead facilitating a ciliary cell fate. However, the post-mitotic or mature RPE was insensitive to the loss, inactivation, or constitutive activation of Ctnnb1. Collectively, our results suggest that Ctnnb1 should be maintained within an optimal range to specify RPE through transcriptional regulation of Wnt target genes in the optic neuroepithelium.
Keywords: β-catenin, ciliary margin, retina, retinal pigment epithelium, Wnt
INTRODUCTION
Three vertebrate optic neural tissues—the neural retina (NR), optic stalk (OS), and retinal pigment epithelium (RPE)—originate in common from a ventral lateral diencephalic neuroepithelial continuum called the optic vesicle (OV) (Chow and Lang, 2001; Heavner and Pevny, 2012). Segregation of the optic neuroepithelial continuum into these three distinct compartments occurs under the influence of external cues that form concentration gradients along the OV (Kim and Kim, 2012). Sonic hedgehog (Shh) diffuses from the ventral medial forebrain and forms a ventral-medial high and dorsal-lateral low gradient along the OV (Zhao et al., 2010). Shh induces the expression of transcription factors, such as ventral anterior homeobox 1 and 2 (Vax1 and -2) and paired homeobox 2 (Pax2), that specify OS fate in the ventral-medial OV compartment (Take-uchi et al., 2003). The dorsal-lateral part of the OV, which is enriched for bone morphogenetic proteins (Bmps), expresses orthodenticle homolog 2 (Otx2) and microphthalmia transcription factor (Mitf) and specifies RPE development against from the adjacent NR compartment, which expresses visual system homeobox 2 (Vsx2) and retina and anterior neural fold homeobox (Rax) (Capowski et al., 2016; Fujimura et al., 2009; Horsford et al., 2005; Westenskow et al., 2009). The loss of these spatially restricted morphogenic cues and transcription factors, therefore, often results in fate transition between optic neuroepithelial compartments (Cardozo et al., 2020).
Wingless/Int1 (Wnt) family proteins—morphogens produced by cells in the mouse mid- and hindbrain—start to affect the optic neuroepithelium during formation of the double layer optic cup (OC) (Fuhrmann, 2008; Machon et al., 2010; Mani et al., 2010). Subsequently, multiple Wnt genes are expressed in the OC in spatially restricted patterns. Wnt2a and Wnt2b are expressed in the cornea and outer ciliary margin (OCM), respectively, whereas Wnt3 is enriched in the retina (Fotaki et al., 2013; Fuhrmann, 2008; Liu et al., 2003; 2006). In addition, Wnt5a is detected at the edge of the inner OC, called the inner ciliary margin (ICM), whereas Wnt5b and Wnt7a are expressed in the lens (Liu et al., 2003). The strongest Wnt signaling activity, which corresponds to the expression of Wnt2 and Wnt3, is found in the peripheral OC area, especially in pigmented cells in the outer layer (Liu et al., 2006). These Wnt ligands induce the expression various genes through stabilization of β-catenin (Ctnnb1), a transcription activator associated with the T cell factor/lymphoid enhancer factor (Tcf/Lef) family transcription factors (Cho and Cepko, 2006; Fujimura et al., 2009; Mosimann et al., 2009; Westenskow et al., 2009). Consistent with the strong Wnt/Ctnnb1 signaling activity in the OCM, previous studies have shown that the RPE fails to develop following loss of the expression of the key determinants, Otx2 and Mitf, in mice lacking Ctnnb1 in the outer OC layer (Westenskow et al., 2009).
The RPE is a typical epithelium, exhibiting a polarized distribution of proteins and strong intercellular junctions (Thumann, 2001). In the mouse RPE, Ctnnb1 was mainly detected in the adherens junction (AJ) (Supplementary Fig. S1), which supports the polarized structure and function of many epithelial cells (Gumbiner, 2005; Le et al., 2021; Perez-Moreno et al., 2003). Moreover, Ctnnb1 dissociates from the AJ and accumulates in the nucleus of the adult mouse RPE during epithelial-to-mesenchymal transition (Kim et al., 2008). Therefore, it is difficult to predict which molecular function of Ctnnb1—component of AJ or key effector of the Wnt signaling pathway—is more critical for acquisition and maintenance of RPE fate.
In this study, we investigated the roles of Ctnnb1 in the developing and mature RPE by genetically dissecting the functions of Ctnnb1. We found that Ctnnb1 is necessary for specifying RPE fate in the optic neuroepithelium. RPE specification by Ctnnb1 was dependent on interactions with the transcription factors, Tcf/Lef and Bcl-9. Interestingly, both loss and constitutive activation of Ctnnb1 resulted in the failure of RPE specification. Constitutive activation of Ctnnb1 converted the RPE to ciliary margin (CM) cells, whereas the inactivation of Ctnnb1 transformed it to the NR. However, neither the activation nor inactivation of Ctnnb1 in the post-mitotic RPE changed RPE structure or function, suggesting that Ctnnb1 is dispensable for RPE maintenance.
MATERIALS AND METHODS
Mice
The mice expressing Ctnnb1flox (Brault et al., 2001), Ctnnb1Δex3 (Harada et al., 1999), Ctnnb1dm (Valenta et al., 2011), and Ctnnb1Y654E (van Veelen et al., 2011) alleles were generated and reported previously. These mice were crossed with Tyrp1-Cre (Mori et al., 2002), Chx10-Cre (Rowan et al., 2004), Mlana-Cre (Aydin and Beermann, 2011), and BEST1-Cre (Lacovelli et al., 2011) mice to delete wild-type Ctnnb1 allele and express functional variants in the outer OC, the inner OC, post-mitotic RPE, and mature RPE, respectively. The ROSA26EYFP (R26EYFP) (JAX stock #006148) and ROSA26tdTomato (R26tdTom; Ai14) (JAX stock #007914) mice were purchased from Jackson Laboratory to monitor the Cre-affected cells in the mice. All experiments were performed under the authorization of the Institutional Animal Care and Use Committee (IACUC) at Korea Advanced Institute of Science and Technology (KAIST) (KA2019-07). All mice used in this study were maintained in a specific pathogen-free facility of KAIST Laboratory Animal Resource Center.
Tissue preparation, histology, immunostaining, and confocal microscopy
The immunostaining of mouse embryonic and adult eyes was done as described in previous reports (Kim et al., 2021; Le et al., 2021). The embryonic heads were isolated from pregnant mice and fixed in phosphate-buffered saline (PBS) solution containing freshly-prepared 4% paraformaldehyde (PFA; Sigma, USA) at 4°C for 2 h. Alternatively, the whole eye balls were isolated from adult mice and fixed in 4% PFA/PBS solution at 4°C for 1 h. The fixed mouse heads and eyes were then transferred to 20% sucrose/PBS solution for 16 h at 4°C. The tissues were then embedded in the Tissue-Tek OCT compound (Sakura Finetek, USA) for freezing those on dry-ice. The frozen tissues were sliced with 10-12 μm thickness by cryostat (CM1805; Leica, Germany) and transferred on the Superfrost Plus Microscope slide (Thermo Fisher Scientific, USA) for H&E staining, which visualizes the nuclei and membranes of the cells in the sections.
For immunostaining, the sections were incubated for 2 h in a blocking solution (10% normal donkey serum in PBS containing 0.2% Triton X-100) at room temperature (RT) for 1 h and subsequently in the solutions containing primary antibodies diluted in the blocking solution at 4°C for 16 h. Antibodies used in this study are listed in Supplementary Table S1. The sections were further stained with Alexa488-, Cy3-, or Alexa647-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, USA) in the blocking solution at RT for 1 h. Fluorescent images were then obtained by confocal microscopy (Flouview FV100 and FV3000; Olympus, Japan) and processed using ImageJ software (NIH).
Cell culture
Human ARPE-19 cells were obtained from the American Tissue Culture Collection (CRL-2302; ATCC, USA) (Ahmado et al., 2011) and maintained in a 37°C humidified incubator with 5% CO2. Cells were cultured in Dulbecco’s modified Eagles’s medium (DMEM) - high glucose (Gibco, 11965; Thermo Fisher Scientific, USA) supplemented with 1% heat inactivated fetal bovine serum (26140079; Gibco - Thermo Fisher Scientific) and 1% penicillin/streptomycin solution (P4333; Sigma-Aldrich).
In situ hybridization (ISH)
Mouse embryonic heads were fixed in freshly-prepared DEPC-PBS solution containing 4% PFA at 4°C for 16 h and were then incubated in DEPC-PBS solution containing 20% sucrose at 4°C for another 16 h. Frozen tissues were sliced at 11 μm thickness for the hybridization with DIG-labeled RNA probes as it was described in a previous report (Balasubramanian et al., 2021). The DIG-labeled Axin2 RNA probes were prepared from pGEM-Axin2 plasmid DNA by in vitro transcription using SP6 RNA polymerase (P1085; Promega, USA). The probes were detected with an alkaline phosphatase-conjugated anti-DIG antibody (11093274910, 1:1,000; Roche, Switzerland) and then visualized by the colorization of NBT/BCIP substrates (11681451001; Roche).
Statistical analyses
Statistical analysis was performed using Prism 7.0 (GraphPad Software, USA). The number of marker positive cells in each image was manually counted. Statistical significance was calculated with Student’s t-test and the error bar in graphs represented the SEM. P values were calculated using an unpaired two-samples t-test. P < 0.05 was considered statistically significant.
RESULTS
Inactivation of the transcription co-activator function of Ctnnb1 transforms the RPE to retina
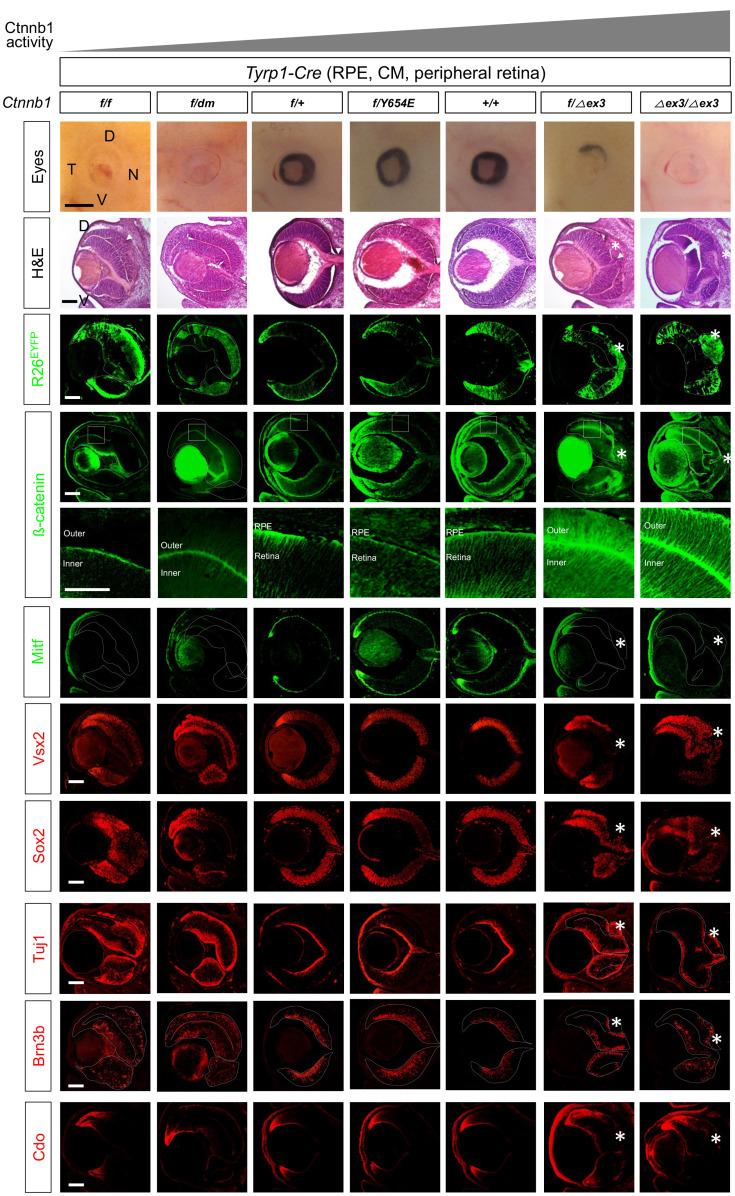
To determine which of Ctnnb1’s functions—mediator of Wnt signaling or component of the AJ—is more critical in mouse RPE development, we replaced mouse Ctnnb1 with variants that selectively disrupt the respective functions. Specifically, we generated mice that express the Ctnnb1 functional variants constitutively (Ctnnb1dm and Ctnnb1Y654E) or upon Cre recombinase-mediated excision (Ctnnb1∆ex3) at the Ctnnb1 gene locus in the RPE cell lineage. We then bred these mice with mice homozygous for the floxed Ctnnb1 gene (Ctnnb1f/f mice), which contains two loxP sequences: one in intron 1 and the other in intron 6 (Brault et al., 2001). In the resulting mice, one copy of the Ctnnb1 gene in the Cre recombinase-affected RPE cell lineage is deleted and the other copy expresses a Ctnnb1 variant.
To abrogate the transcription-activator function of Ctnnb1 in RPE lineages, we first bred Ctnnb1 double transcription mutant (Ctnnb1dm) mice with Ctnnb1f/f;Tyrp1-Cre mice to obtain Ctnnb1f/dm;Tyrp1-Cre mice. In the Ctnnb1dm mutant, aspartate 164 (D164), which is necessary for the interaction with Bcl9, is changed to alanine, and the C-terminal region, which is responsible for interaction with Tcf/Lef transcription factors, is deleted (Mori et al., 2002; Valenta et al., 2011). Therefore, in Ctnnb1f/dm;Tyrp1-Cre mice, only the Ctnnb1dm mutant is expressed in Cre-affected RPE, CM, and retinal subpopulations, while cells in the rest of body express both wild-type Ctnnb1 and the Ctnnb1dm mutant. However, Ctnnb1dm/dm homozygous mutant mice could not survive to the birth, as it was reported previously (Valenta et al., 2011).
In consistent with the idea that Ctnnb1 supports development of the RPE by inducing the expression of RPE-specifying genes, the outer OC layers of Ctnnb1f/dm;Tyrp1-Cre mice were depigmented and lost expression of Mitf, an RPE marker (Fig. 1, the second leftmost column). Instead, these cells expressed the retinal markers, Vsx2 and Sox2, which are expressed in retinal progenitor cells (RPCs), and tubulin β-III (detected with the Tuj1 antibody), which are expressed in post-mitotic neurons (PMNs) (Fig. 1, the second leftmost column). They also expressed Brn3b, which is a marker of RGCs (retinal ganglion cells). These phenotypes are almost identical to those of Ctnnb1f/f;Tyrp1-Cre mice (Fig. 1, the leftmost column), with the exception of retinal rosettes, which were not seen only in the Ctnnb1f/dm;Tyrp1-Cre mouse retina (Fig. 1, arrowhead in the H&E images at the leftmost column points the rosette). These results therefore suggest that Ctnnb1 supports RPE development by inducing the expression of genes necessary for the specification and/or maintenance of the RPE.
Fig. 1. Transcription regulator function of Ctnnb1 is necessary for RPE development.
Pictures in two top rows are embryonic day 14.5 (E14.5) mouse eyes (side view; top row) and H&E staining images of the eye sections (second row), which have wild-type (+), floxed exon 2-6 (f), dm, Y654E, and floxed exon 3 (∆ex3) alleles of Ctnnb1 in Tyrp1-Cre-affected retinal pigment epithelium (RPE), outer ciliary margin (OCM), inner ciliary margin (ICM) and peripheral retinal subpopulations. D, dorsal; V, ventral; N, nasal; T, temporal. Expression of R26EYFP Cre recombinase reporter (third row) and Ctnnb1 (fourth and fifth rows) in the eye sections were examined by immunostaining of EYFP and Ctnnb1, respectively. Images in the fifth row are magnified version of the boxed areas in the fourth row. The eye sections were also immunostained with the antibodies detecting cell type-specific markers in the following. Mitf, RPE and OCM; Vsx2, medial and proximal ICM and retinal progenitor cell (RPC); Sox2, proximal ICM and RPC, anti-tubulin βIII (Tuj1), PMNs (post-mitotic neurons); Brn3b, RGCs (retinal ganglion cells); Cdo, ICM. Dotted lines are the edges of the OC. Scale bars = 500 μm (first row), 50 μm (fifth row), and 100 μm (the rest rows). Asterisks, ectopic CM in RPE-OS border; arrowheads, retinal rosette.
RPE development is independent of the phosphorylation status of Ctnnb1 residue Y654
It has been shown that junctional dynamics of the epithelium are sensitive to the phosphorylation status of Ctnnb1 (Daugherty and Gottardi, 2007). In particular, the phosphorylation of tyrosine residues, which are targets of intracellular tyrosine kinases, including Src (Roura et al., 1999), Abl (Rhee et al., 2002), Fyn, Fer and cMet (Piedra et al., 2003), was found to result in the dissociation of Ctnnb1 from the AJ. Phosphorylation of Ctnnb1 is also significantly elevated in the mouse RPE, which exhibits a decrease in junctional Ctnnb1 but shows an increase in nuclear Ctnnb1 that triggers the mesenchymal transition (Kim et al., 2008). The phosphorylation status of Y654 was found to be critical for embryonic development, as evidenced by the fact that Ctnnb1Y654E/Y654E mice die neonatally owing to multiple morphological defects in head structures (van Veelen et al., 2011). We also found that the Y654E mutation disrupted the junctional association of Ctnnb1 in ARPE-19 cells, causing Ctnnb1 (Y654E) protein to adopt a diffuse intracellular distribution (Supplementary Fig. S2). Conversely, mutating Y654 to a phosphorylation-defective phenylalanine (Y654F) enhanced membrane localization of Ctnnb1 (Supplementary Fig. S2). Accordingly, we investigated the roles of Ctnnb1 as a junctional component in RPE development and maintenance in Ctnnb1f/Y654E;Tyrp1-Cre mice, obtained by breeding Ctnnb1f/f;Tyrp1-Cre mice with Ctnnb1Y654E/+ mice. However, this genetic manipulation did not affect development of the RPE, which showed appropriate basal and lateral expression of Ctnnb1Y654E proteins (Fig. 1, the center column). These results therefore suggest that phosphorylation of Y654 alone is not sufficient to impair the junctional localization of Ctnnb1 in the optic neuroepithelium and RPE.
RPE maintenance requires Wnt/Ctnnb1 signaling activity within an appropriate range
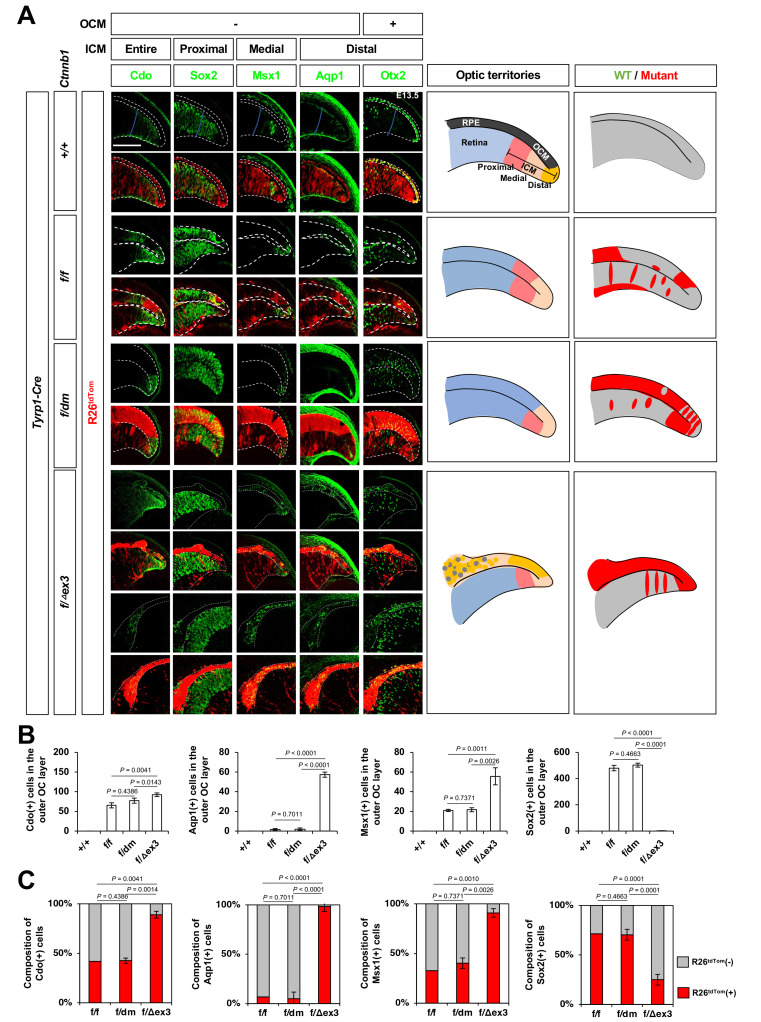
Wnt/Ctnnb1 signaling activity, determined by examining the expression of its target Axin2 by ISH, was detected in the distal RPE layer, which contains proliferating pigmented cells and is defined as the OCM, but was not detectable in the central RPE or retina in the central optic cup (Supplementary Fig. S3). The Axin2 ISH signals disappeared completely in the eyes of Ctnnb1f/f;Tyrp1-Cre and Ctnnb1f/dm;Tyrp1-Cre mice (Supplementary Fig. S3). These results therefore suggest that the Wnt/Ctnnb1 pathway is active in RPE progenitor cells in the OCM.
We, thus, tested the intriguing hypothesis that RPE progenitor cells could expand persistently if Wnt/Ctnnb1 signaling is activated constitutively. To test this, we replaced Ctnnb1 in the mouse RPE lineage cells with the Ctnnb1∆ex3 mutant, which is expressed Cre recombinase-mediated deletion of exon 3 (ex3) encoding sequences containing serine 9 (Ser9), a phosphorylation site for glycogen synthase kinase 3 (Gsk3) (Harada et al., 1999). The loss of this site causes the Ctnnb1∆ex3 protein to become resistant to ubiquitin-dependent degradation, which is triggered by the phosphorylation of Ctnnb1 at Ser9, allowing the protein to accumulate in cells even in the absence of Wnt-induced Gsk3 inhibition (Harada et al., 1999). The Axin2 mRNA expression expanded into the entire outer OC of Ctnnb1f/∆ex3;Tyrp1-Cre mice (Supplementary Fig. S3). The results suggest that Ctnnb1 was activated constitutively to induce its target genes ectopically in the RPE, which lost pigments and overproliferated.
Despite the fact that the ∆ex3 mutation has the opposite effect from the dm mutation on Ctnnb1 activity, the eye phenotypes of Ctnnb1f/∆ex3;Tyrp1-Cre mouse embryos were similar to those of Ctnnb1f/dm;Tyrp1-Cre mouse embryos (Fig. 1). However, in contrast to Ctnnb1f/dm;Tyrp1-Cre mouse eyes, the outer OC layer cells in Ctnnb1f/∆ex3;Tyrp1-Cre and Ctnnb1∆ex3/∆ex3;Tyrp1-Cre mice were not transformed into the retinal cells, which express Vsx2-, Sox2-, or Brn3b. Instead, they were found to express the ICM markers, Cdo, Msx1, and Aqp1 (Figs. 1 [bottom row], 2A, and 2B).
Fig. 2. Ctnnb1 facilitates distal ciliary margin (CM) cell fate in the optic neuroepithelium.
(A) Sections of embryonic day 13.5 (E13.5) Ctnnb1+/+;Tyrp1-Cre, Ctnnb1f/f;Tyrp1-Cre, Ctnnb1f/dm;Tyrp1-Cre, and Ctnnb1f/Δex3;Tyrp1-Cre mouse eyes were stained with the antibodies against Cdo, Sox2, Msx1, Aqp1, and Otx2. Optic neuroepithelial continuum of the retina (inner) and RPE (outer) are marked by the dotted lines. Schematic diagrams depict the peripheral optic cup areas. Scale bar = 50 μm. (B) The cells identified by each cell type-specific marker were counted and the numbers are shown in the graphs. (C) The R26tdTom Cre reporter-expressing cell populations in the marker-positive cells are shown in the graphs. Values in the y-axis are averages and error bars denote the SEM. n = 5, 3 independent litters. OCM, outer ciliary margin; ICM, inner ciliary margin; RPE, retinal pigment epithelium; WT, wild type; OC, optic cup.
The ectopic ICM cells were also detectable in the depigmented outer OC layers of Ctnnb1f/f;Tyrp1-Cre and Ctnnb1f/dm;Tyrp1-Cre mouse eyes (Figs. 1, 2A, and 2B), in which Ctnnb1 were absent or inactivated, respectively. However, according to the mosaic expression pattern of ROSA26EYFP (R26EYFP) and ROSA26tdTomato (R26tdTom) Cre reporters in the mouse eyes (Figs. 1A [third row], 2A, and 2C), the cells in the outer OC layers of Ctnnb1f/f;Tyrp1-Cre, Ctnnb1f/dm;Tyrp1-Cre, and Ctnnb1f/∆ex3;Tyrp1-Cre mouse eyes were mixed populations of Cre-unaffected wild-type and Cre-affected mutant cells. The majority of Cdo-positive ICM cells in the outer OC layers of Ctnnb1f/f;Tyrp1-Cre and Ctnnb1f/dm;Tyrp1-Cre mice were R26tdTom-negative (Figs. 2A and 2C), suggesting that they express wild-type Ctnnb1. In contrast, the ICM cells in the outer OC layer of Ctnnb1f/∆ex3;Tyrp1-Cre mice were positive to R26tdTom Cre reporter (Figs. 2A and 2C), indicating that those express Ctnnb1∆ex3 instead of wild-type Ctnnb1. These results therefore suggest that Ctnnb1-dependent gene expression in the outer OC layer facilitates CM fate but suppresses the differentiation to the RPE in cell autonomous manners.
Suppression of Wnt/Ctnnb1 signaling activity is necessary for retinal development
We next tested whether a loss or gain of Ctnnb1 function also converts the fates of retina to RPE or CM. To this end, we deleted Ctnnb1 or expressed Ctnnb1 functional variants in Vsx2/Chx10-positive RPCs and medial-proximal ICM populations using Chx10-Cre (Rowan et al., 2004). We found that Ctnnb1 loss (Ctnnb1f/f;Chx10-Cre) or inactivation (Ctnnb1f/dm;Chx10-Cre) in these cells and their descendent cells did not change the expression of retinal (Vsx2, Sox2, Brn3b, and Tubb3) or RPE (Mitf) markers (Supplementary Fig. S4). Multiple rosettes were observed in Ctnnb1f/f;Chx10-Cre mouse retinas but not in Ctnnb1f/dm;Chx10-Cre mouse retinas, suggesting that Ctnnb1 plays a critical role in the maintenance of retinal structural integrity but is dispensable for retinal fate acquisition and/or maintenance. However, no remarkable changes were observed in Ctnnb1f/Y654E;Chx10-Cre mice (Supplementary Fig. S4).
Instead, the rosettes were found in Ctnnb1f/∆ex3;Chx10-Cre and Ctnnb1∆ex3/∆ex3;Chx10-Cre mouse retinas (Supplementary Fig. S4, two rightmost columns). The majority of retinal cells in these mice did not express the RPC markers, Vsx2 and Sox2, or the PMN markers, Brn3b and Tubb3. Instead, cells expressing the CM marker, Cdo, were increased in the retinal area (Supplementary Fig. S4). This was reminiscent to the results of Ctnnb1f/∆ex3;Tyrp1-Cre and Ctnnb1∆ex3/∆ex3;Tyrp1-Cre mice, in which the cells in the RPE layer were transformed into the ICM cells (Figs. 1 and 2). These results therefore suggest that hyperactivation of Ctnnb1 facilitates CM fate acquisition commonly in the inner and outer OC layers. Consequently, this interfered with the development of RPE and retina in the inner and outer optic cup layers, respectively, resulting in the microphthalmia of Ctnnb1∆ex3 expressing mouse eyes (Fig. 1, Supplementary Fig. S4, two rightmost columns).
Ctnnb1 is dispensable for RPE maintenance
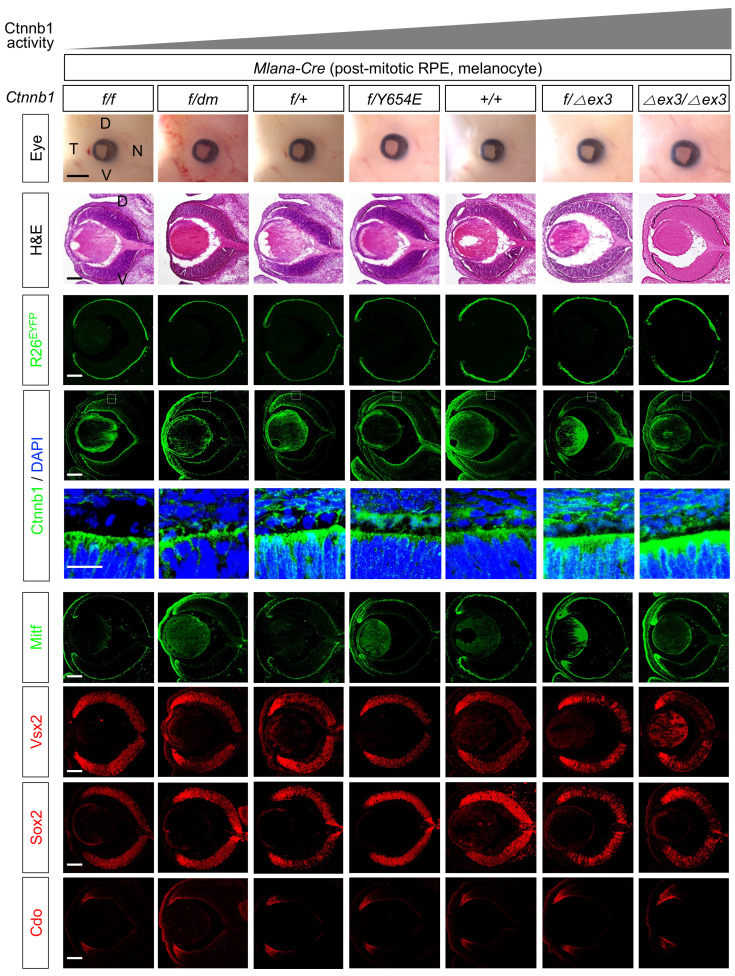
Next, to determine whether Ctnnb1 is critical for the maintenance of RPE, as well as the specification, we deleted Ctnnb1 or expressed Ctnnb1 functional variants in the post-mitotic RPE using Mlana1-Cre (Aydin and Beermann, 2011). The Cre-mediated deletion of Ctnnb1 in Ctnnb1f/f;Mlana1-Cre mice, however, did not significantly affect eye development (Fig. 3, top row). Ctnnb1f/dm;Mlana1-Cre, Ctnnb1f/654E;Mlana1-Cre, Ctnnb1f/∆ex3;Tyrp1-Cre, and Ctnnb1∆ex3/∆ex3;Tyrp1-Cre mice also exhibited a normal eye morphology (Fig. 3, top row), although Ctnnb1 proteins were more abundant in these mouse RPE than in the wild-type RPE of Ctnnb1+/+;Mlana1-Cre mice (Fig. 3, fourth row). In contrast, loss of Ctnnb1 (Ctnnb1f/f) or impaired target gene expression (Ctnnb1f/dm) resulted in changes in coat color (Supplementary Fig. S5), suggesting a critical role for Ctnnb1 in melanocyte development and maintenance.
Fig. 3. Ctnnb1 is dispensable for the post-mitotic retinal pigment epithelium (RPE).
Pictures of the eyes and H&E staining images of the eye sections of embryonic day 14.5 (E14.5) mice, which express wild-type and functional variants of Ctnnb1, are provided in the first and second rows, respectively. Expression of R26EYFP Cre recombinase reporter (third row) and Ctnnb1 (fourth and fifth rows) in the mouse eyes were examined by immunostaining of EYFP and Ctnnb1, respectively. Images in the fifth row show magnified versions of the boxed areas in the fourth row. The eye sections were also immunostained with the antibodies against specific markers to identify the distribution of corresponding cell types. D, dorsal; V, ventral; N, nasal; T, temporal. Scale bars = 500 μm (first row), 50 μm (fifth row), and 100 μm (the rest rows).
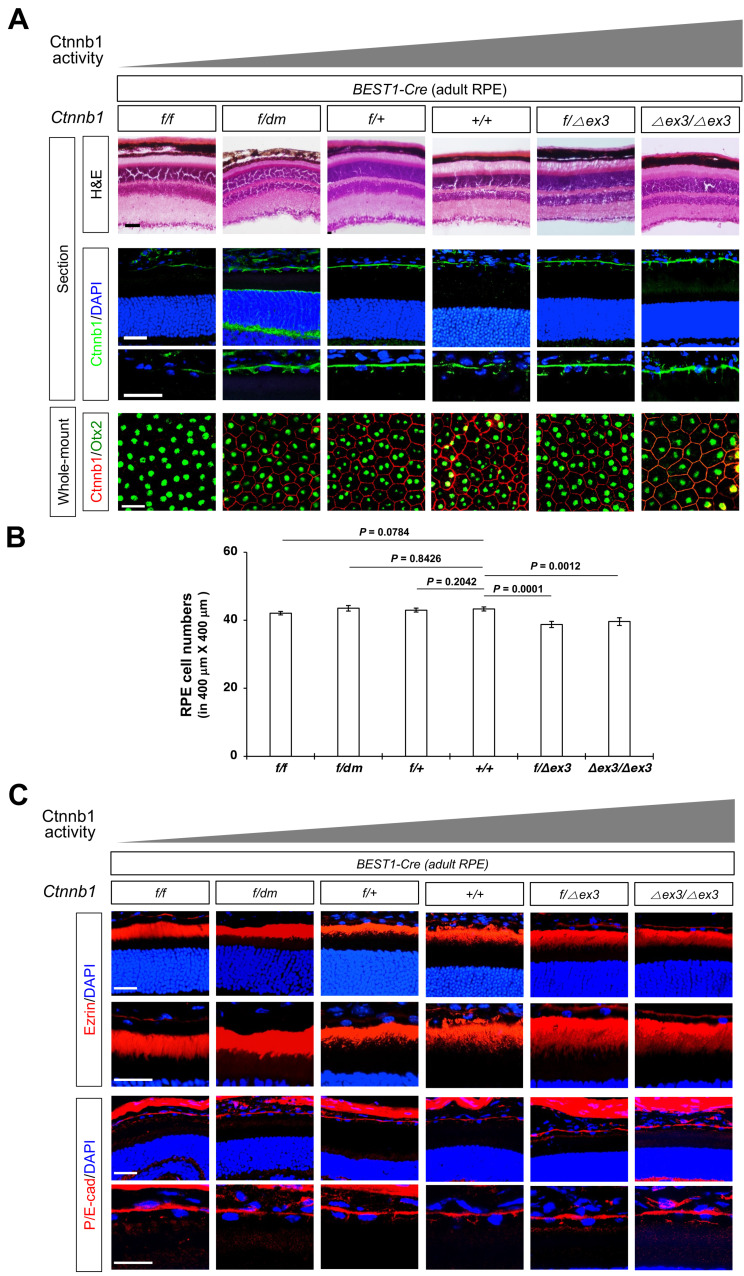
We also investigated the roles of Ctnnb1 in adult mouse RPE by deleting or expressing functional variants using BEST1-Cre, which is expressed in the mouse RPE after P14 (Lacovelli et al., 2011). However, we detected no remarkable anatomical changes in the RPE of Ctnnb1f/f;BEST1-Cre, Ctnnb1f/dm;BEST1-Cre, Ctnnb1f/∆ex3;BEST1-Cre, or Ctnnb1∆ex3/∆ex3;BEST1-Cre mice compared with that of Ctnnb1+/+;BEST1-Cre mice (Figs. 4A and 4B). The apico-basal polarity of the RPE, which is determined by the distributions of an apical RPE marker ezrin and a basolateral RPE marker E-cadherin (E-cad), were also well maintained (Fig. 4C). These results imply that Ctnnb1 is dispensable for the maintenance of the mouse RPE but is necessary for its specification.
Fig. 4. Ctnnb1 is dispensable for mature retinal pigment epithelium (RPE).
(A) Structures of P60 mouse eyes, which express wild-type and functional variants of Ctnnb1 in BEST1-Cre affected mature RPE, are visualized by H&E staining (top row). Ctnnb1 expression was examined by immunostaining of the mouse eye sections (second and third row). Intercellular junctions and the nuclei of RPE in the whole-mount mouse eye cups were visualized by immunostaining against the antibodies against Ctnnb1 and Otx2, respectively (fourth row). (B) Numbers of Otx2-positive RPE cells in the indicated areas of whole-mount eye cups were counted and shown in the graph. Values in the y-axis are averages and error bard denote SEM. n ≥ 22 from 6 mouse eyes (3 independent litters). (C) Distribution of the microvilli and AJ in P60 mouse RPE were investigated by immunostaining of Ezrin and P/E-cadherin, respectively. Images in the second and fourth rows show magnified version of RPE in first and third rows, respectively. Scale bars = 50 μm (A and C).
DISCUSSION
Spatial segregation of the neuroepithelial continuum is necessary for the development of various tissues comprising the central nervous system. The process is known to be regulated by morphogens, which form concentration gradients that act on the neuroepithelial continuum. In E8.5 mouse embryos, Wnt/Ctnnb1 signaling activity has been identified in the diencephalic area, where the OV extends laterally (Maretto et al., 2003). Wnt/Ctnnb1 signaling was found to be essential for OV formation, as shown by the failure of eyes to develop in mice lacking Ctnnb1 in forebrain areas (Hagglund et al., 2013). Then, Wnt/Ctnnb1 signaling activity is restricted to the dorsal OV (Fuhrmann, 2008; Machon et al., 2010; Mani et al., 2010), a primitive RPE that enwraps the retina during invagination of the OV to form the double-layer OC. Ctnnb1 in the dorsal OV was previously reported to be necessary for RPE development, a conclusion supported by the observation that the Ctnnb1-deficient RPE transforms into the retina (Fujimura et al., 2009; Westenskow et al., 2009). However, how Ctnnb1 is involved in the specification and/or maintenance RPE against default retinal fate acquisition has not been clearly established. In this study, we show that Ctnnb1 is necessary for the specification, but not the maintenance, of RPE cell fate in the outer OC layer (Figs. 1, 3, and 4).
Our results further show that eye development requires an appropriate dosage of Ctnnb1. Expression of the constitutively active Ctnnb1∆ex3 mutant in the dorsal OV resulted in conversion of the RPE to a neuroepithelial cell population found in the CM, whereas the loss of Ctnnb1 resulted in transformation of the RPE to a retina (Fig. 1). Constitutive activation of Ctnnb1 also interfered with formation of the RPE-OS border structure in the Ctnnb1∆ex3 mutant mice. Ectopic cells in the RPE-OS border did not develop into the RPE, which expresses Mitf, or the OS, which expresses Pax2 (Fig. 1, Supplementary Fig. S6, asterisk). Instead, these cells exhibited characteristics of ICM cells, which express Cdo (Fig. 1, two rightmost columns). These results suggest that the divergent development of CM populations into the retina and RPE requires the suppression of Wnt/Ctnnb1 signaling outside of the CM. Otherwise, the development and growth of double-layered OC is impaired (Fig. 1, Supplementary Fig. S4, two rightmost columns).
The spatial restriction of ICM cells in the peripheral OC area (i.e., RPE-NR border) suggests the presence of factors that suppress Wnt/Ctnnb1 signaling in the central OC. Secreted frizzled receptor 1 (Sfrp1) and 2 (Sfrp2) are expressed in the peripheral and central retina of E13.5 mouse embryos, respectively, whereas Sfrp3 is expressed in the RPE (Liu et al., 2003). Thus, these Wnt antagonists might interfere with the binding of Wnt ligands expressed in the retina (Wnt3, Wnt5a, Wnt5b, and Wnt7b), RPE (Wnt13), OCM (Wnt13), ICM (Wnt3 and Wnt5a), lens (Wnt3, Wnt5a, and Wnt5b), and cornea (Wnt2, Wnt3, Wnt4, and Wnt6) to their receptors, such as Mfz-3 and Mfz-7 (Liu et al., 2003). However, CM fate is not enhanced in mice lacking Sfrp1 and Sfrp2, which exhibited failure of CB specification in the peripheral OC (Esteve et al., 2011). These results suggest that Sfrp1 and Sfrp2 instead support Wnt/Ctnnb1–dependent CM and RPE fate specification. The Wnt antagonists, Dickkopf (Dkk1) and Dkk3, are present in the ICM and retina, respectively (Lieven and Ruther, 2011; Sato et al., 2007). Heterozygous loss of Dkk1 results in a malformation of the eye in which the border between the retina and OS is missing and the dorsal OC is expanded (Lieven and Ruther, 2011). However, peripheral patterning of the OC is not significantly affected in these mice. Thus, the critical roles of Dkk1 in the CM should be investigated further using an OC-specific loss- or gain-of-function strategy.
The AJs, which are supported by Ctnnb1, have been identified to be necessary for the maintenance of polarized structure of RPE (Thumann, 2001). Therefore, it was surprising that Ctnnb1 is dispensable for the maintenance of RPE (Figs. 3 and 4). Our results suggest that other catenins, such as δ-catenin (Ctnnd1)/p120-catenin and γ-catenin (Ctnng)/Plakoglobin, might compensate Ctnnb1 in the mouse RPE. This hypothesis should be tested in future studies by inactivating multiple catenin genes simultaneously in RPE.
Activation of Wnt/Ctnnb1 pathway has been also shown to disrupt RPE structure via the epithelial-to-mesenchyme transition (EMT) of mature RPE (Han et al., 2015; Kim et al., 2008; Zhou et al., 2020). However, the expression of constitutively active Ctnnb1∆ex3 did not result in the EMT of RPE in Ctnnb1f/∆ex3;BEST1-Cre and Ctnnb1∆ex3/∆ex3;BEST1-Cre mice (Fig. 4). Furthermore, in contrast to the observations in the previously reported pathological conditions that mobilized Ctnnb1 from the AJ to the nucleus in RPE (Han et al., 2015; Kim et al., 2008), the mutation did not eliminate Ctnnb1 from the AJs, while it could elevate Ctnnb1 in the cytoplasm and nucleus (Fig. 4). Therefore, those previously reported pathological changes of RPE should not be solely resulted from the activation of Ctnnb1 but might be combined outputs of multiple intracellular signaling pathways, including PI3K-Akt and Ras-MAPK pathways. Therefore, it would be necessary to develop the methods that control multiple pathways to suppress the EMT in RPE pathogenesis.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by National Research Foundation of Korea (NRF) grants (NRF-2022R1A2C3003589; NRF-2018R1A5A1024261) funded by Korean Ministry of Science and ICT (MSIT) and the International Collaboration Initiative grant (KAIST-N11210255) supported by KAIST, South Korea.
Footnotes
AUTHOR CONTRIBUTIONS
J.M.K., K.W.M., and Y.J.K. performed experiments. R.S. and K.B. provided the research materials. J.W.K. conceived and supervised the experiments, wrote the manuscript, and secured funding.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahmado A., Carr A.J., Vugler A.A., Semo M., Gias C., Lawrence J.M., Chen L.L., Chen F.K., Turowski P., da Cruz L., et al. Induction of differentiation by pyruvate and DMEM in the human retinal pigment epithelium cell line ARPE-19. Invest. Ophthalmol. Vis. Sci. 2011;52:7148–7159. doi: 10.1167/iovs.10-6374. [DOI] [PubMed] [Google Scholar]
- Aydin I.T., Beermann F. A mart-1::Cre transgenic line induces recombination in melanocytes and retinal pigment epithelium. Genesis. 2011;49:403–409. doi: 10.1002/dvg.20725. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R., Min X., Quinn P.M.J., Giudice Q.L., Tao C., Polanco K., Makrides N., Peregrin J., Bouaziz M., Mao Y., et al. Phase transition specified by a binary code patterns the vertebrate eye cup. Sci. Adv. 2021;7:eabj9846. doi: 10.1126/sciadv.abj9846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D.H., McMahon A.P., Sommer L., Boussadia O., Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Capowski E.E., Wright L.S., Liang K., Phillips M.J., Wallace K., Petelinsek A., Hagstrom A., Pinilla I., Borys K., Lien J., et al. Regulation of WNT signaling by VSX2 during optic vesicle patterning in human induced pluripotent stem cells. Stem Cells. 2016;34:2625–2634. doi: 10.1002/stem.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo M.J., Almuedo-Castillo M., Bovolenta P. Patterning the vertebrate retina with morphogenetic signaling pathways. Neuroscientist. 2020;26:185–196. doi: 10.1177/1073858419874016. [DOI] [PubMed] [Google Scholar]
- Cho S.H., Cepko C.L. Wnt2b/beta-catenin-mediated canonical Wnt signaling determines the peripheral fates of the chick eye. Development. 2006;133:3167–3177. doi: 10.1242/dev.02474. [DOI] [PubMed] [Google Scholar]
- Chow R.L., Lang R.A. Early eye development in vertebrates. Annu. Rev. Cell Dev. Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- Daugherty R.L., Gottardi C.J. Phospho-regulation of Beta-catenin adhesion and signaling functions. Physiology (Bethesda) 2007;22:303–309. doi: 10.1152/physiol.00020.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve P., Sandonis A., Ibanez C., Shimono A., Guerrero I., Bovolenta P. Secreted frizzled-related proteins are required for Wnt/beta-catenin signalling activation in the vertebrate optic cup. Development. 2011;138:4179–4184. doi: 10.1242/dev.065839. [DOI] [PubMed] [Google Scholar]
- Fotaki V., Smith R., Pratt T., Price D.J. Foxg1 is required to limit the formation of ciliary margin tissue and Wnt/beta-catenin signalling in the developing nasal retina of the mouse. Dev. Biol. 2013;380:299–313. doi: 10.1016/j.ydbio.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann S. Wnt signaling in eye organogenesis. Organogenesis. 2008;4:60–67. doi: 10.4161/org.4.2.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura N., Taketo M.M., Mori M., Korinek V., Kozmik Z. Spatial and temporal regulation of Wnt/beta-catenin signaling is essential for development of the retinal pigment epithelium. Dev. Biol. 2009;334:31–45. doi: 10.1016/j.ydbio.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Gumbiner B.M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Hagglund A.C., Berghard A., Carlsson L. Canonical Wnt/beta-catenin signalling is essential for optic cup formation. PLoS One. 2013;8:e81158. doi: 10.1371/journal.pone.0081158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.W., Lyu J., Park Y.J., Jang S.Y., Park T.K. Wnt/beta-catenin signaling mediates regeneration of retinal pigment epithelium after laser photocoagulation in mouse eye. Invest. Ophthalmol. Vis. Sci. 2015;56:8314–8324. doi: 10.1167/iovs.15-18359. [DOI] [PubMed] [Google Scholar]
- Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M., Taketo M.M. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner W., Pevny L. Eye development and retinogenesis. Cold Spring Harb. Perspect. Biol. 2012;4:a008391. doi: 10.1101/cshperspect.a008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsford D.J., Nguyen M.T., Sellar G.C., Kothary R., Arnheiter H., McInnes R.R. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development. 2005;132:177–187. doi: 10.1242/dev.01571. [DOI] [PubMed] [Google Scholar]
- Kim H.T., Kim J.W. Compartmentalization of vertebrate optic neuroephithelium: external cues and transcription factors. Mol. Cells. 2012;33:317–324. doi: 10.1007/s10059-012-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.W., Kang K.H., Burrola P., Mak T.W., Lemke G. Retinal degeneration triggered by inactivation of PTEN in the retinal pigment epithelium. Genes Dev. 2008;22:3147–3157. doi: 10.1101/gad.1700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Park S., Ha T., Kim S., Lim S., You H., Kim J.W. Retinoid metabolism in the degeneration of Pten-deficient mouse retinal pigment epithelium. Mol. Cells. 2021;44:613–622. doi: 10.14348/molcells.2021.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacovelli J., Zhao C., Wolkow N., Veldman P., Gollomp K., Ojha P., Lukinova N., King A., Feiner L., Esumi N., et al. Generation of Cre transgenic mice with postnatal RPE-specific ocular expression. Invest. Ophthalmol. Vis. Sci. 2011;52:1378–1383. doi: 10.1167/iovs.10-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le D., Lim S., Min K.W., Park J.W., Kim Y., Ha T., Moon K.H., Wagner K.U., Kim J.W. Tsg101 is necessary for the establishment and maintenance of mouse retinal pigment epithelial cell polarity. Mol. Cells. 2021;44:168–178. doi: 10.14348/molcells.2021.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieven O., Ruther U. The Dkk1 dose is critical for eye development. Dev. Biol. 2011;355:124–137. doi: 10.1016/j.ydbio.2011.04.023. [DOI] [PubMed] [Google Scholar]
- Liu H., Mohamed O., Dufort D., Wallace V.A. Characterization of Wnt signaling components and activation of the Wnt canonical pathway in the murine retina. Dev. Dyn. 2003;227:323–334. doi: 10.1002/dvdy.10315. [DOI] [PubMed] [Google Scholar]
- Liu H., Thurig S., Mohamed O., Dufort D., Wallace V.A. Mapping canonical Wnt signaling in the developing and adult retina. Invest. Ophthalmol. Vis. Sci. 2006;47:5088–5097. doi: 10.1167/iovs.06-0403. [DOI] [PubMed] [Google Scholar]
- Machon O., Kreslova J., Ruzickova J., Vacik T., Klimova L., Fujimura N., Lachova J., Kozmik Z. Lens morphogenesis is dependent on Pax6-mediated inhibition of the canonical Wnt/beta-catenin signaling in the lens surface ectoderm. Genesis. 2010;48:86–95. doi: 10.1002/dvg.20583. [DOI] [PubMed] [Google Scholar]
- Mani P., Jarrell A., Myers J., Atit R. Visualizing canonical Wnt signaling during mouse craniofacial development. Dev. Dyn. 2010;239:354–363. doi: 10.1002/dvdy.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S., Cordenonsi M., Dupont S., Braghetta P., Broccoli V., Hassan A.B., Volpin D., Bressan G.M., Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Metzger D., Garnier J.M., Chambon P., Mark M. Site-specific somatic mutagenesis in the retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 2002;43:1384–1388. [PubMed] [Google Scholar]
- Mosimann C., Hausmann G., Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M., Jamora C., Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/S0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- Piedra J., Miravet S., Castano J., Palmer H.G., Heisterkamp N., Garcia, de Herreros A., Dunach M. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate beta-catenin Tyr-142 phosphorylation and beta-catenin-alpha-catenin interaction. Mol. Cell. Biol. 2003;23:2287–2297. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J., Mahfooz N.S., Arregui C., Lilien J., Balsamo J., VanBerkum M.F. Activation of the repulsive receptor Roundabout inhibits N-cadherin-mediated cell adhesion. Nat. Cell Biol. 2002;4:798–805. doi: 10.1038/ncb858. [DOI] [PubMed] [Google Scholar]
- Roura S., Miravet S., Piedra J., Garcia, de Herreros A., Dunach M. Regulation of E-cadherin/Catenin association by tyrosine phosphorylation. J. Biol. Chem. 1999;274:36734–36740. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- Rowan S., Chen C.M., Young T.L., Fisher D.E., Cepko C.L. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–5152. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- Sato S., Inoue T., Terada K., Matsuo I., Aizawa S., Tano Y., Fujikado T., Furukawa T. Dkk3-Cre BAC transgenic mouse line: a tool for highly efficient gene deletion in retinal progenitor cells. Genesis. 2007;45:502–507. doi: 10.1002/dvg.20318. [DOI] [PubMed] [Google Scholar]
- Take-uchi M., Clarke J.D., Wilson S.W. Hedgehog signalling maintains the optic stalk-retinal interface through the regulation of Vax gene activity. Development. 2003;130:955–968. doi: 10.1242/dev.00305. [DOI] [PubMed] [Google Scholar]
- Thumann G. Development and cellular functions of the iris pigment epithelium. Surv. Ophthalmol. 2001;45:345–354. doi: 10.1016/S0039-6257(00)00195-8. [DOI] [PubMed] [Google Scholar]
- Valenta T., Gay M., Steiner S., Draganova K., Zemke M., Hoffmans R., Cinelli P., Aguet M., Sommer L., Basler K. Probing transcription-specific outputs of beta-catenin in vivo. Genes Dev. 2011;25:2631–2643. doi: 10.1101/gad.181289.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen W., Le N.H., Helvensteijn W., Blonden L., Theeuwes M., Bakker E.R., Franken P.F., van Gurp L., Meijlink F., van der Valk M.A., et al. beta-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut. 2011;60:1204–1212. doi: 10.1136/gut.2010.233460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenskow P., Piccolo S., Fuhrmann S. Beta-catenin controls differentiation of the retinal pigment epithelium in the mouse optic cup by regulating Mitf and Otx2 expression. Development. 2009;136:2505–2510. doi: 10.1242/dev.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Saitsu H., Sun X., Shiota K., Ishibashi M. Sonic hedgehog is involved in formation of the ventral optic cup by limiting Bmp4 expression to the dorsal domain. Mech. Dev. 2010;127:62–72. doi: 10.1016/j.mod.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Zhou M., Geathers J.S., Grillo S.L., Weber S.R., Wang W., Zhao Y., Sundstrom J.M. Role of epithelial-mesenchymal transition in retinal pigment epithelium dysfunction. Front. Cell Dev. Biol. 2020;8:501. doi: 10.3389/fcell.2020.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.