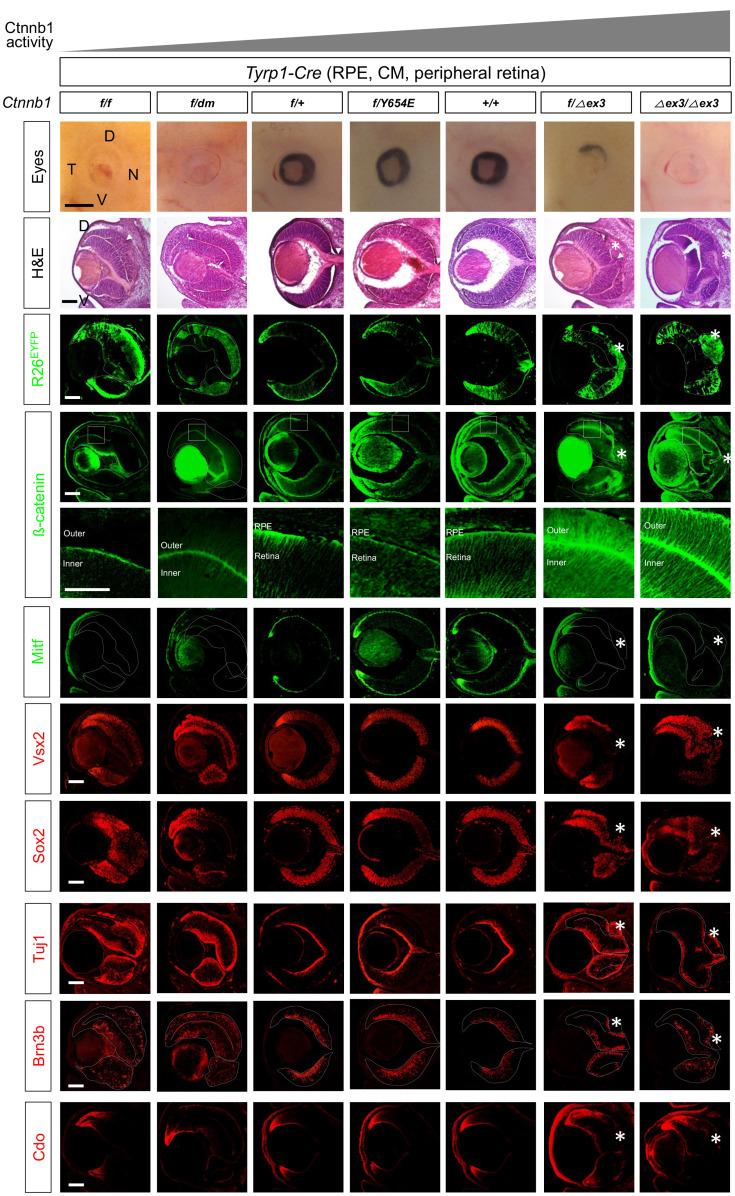
Fig. 1. Transcription regulator function of Ctnnb1 is necessary for RPE development.
Pictures in two top rows are embryonic day 14.5 (E14.5) mouse eyes (side view; top row) and H&E staining images of the eye sections (second row), which have wild-type (+), floxed exon 2-6 (f), dm, Y654E, and floxed exon 3 (∆ex3) alleles of Ctnnb1 in Tyrp1-Cre-affected retinal pigment epithelium (RPE), outer ciliary margin (OCM), inner ciliary margin (ICM) and peripheral retinal subpopulations. D, dorsal; V, ventral; N, nasal; T, temporal. Expression of R26EYFP Cre recombinase reporter (third row) and Ctnnb1 (fourth and fifth rows) in the eye sections were examined by immunostaining of EYFP and Ctnnb1, respectively. Images in the fifth row are magnified version of the boxed areas in the fourth row. The eye sections were also immunostained with the antibodies detecting cell type-specific markers in the following. Mitf, RPE and OCM; Vsx2, medial and proximal ICM and retinal progenitor cell (RPC); Sox2, proximal ICM and RPC, anti-tubulin βIII (Tuj1), PMNs (post-mitotic neurons); Brn3b, RGCs (retinal ganglion cells); Cdo, ICM. Dotted lines are the edges of the OC. Scale bars = 500 μm (first row), 50 μm (fifth row), and 100 μm (the rest rows). Asterisks, ectopic CM in RPE-OS border; arrowheads, retinal rosette.