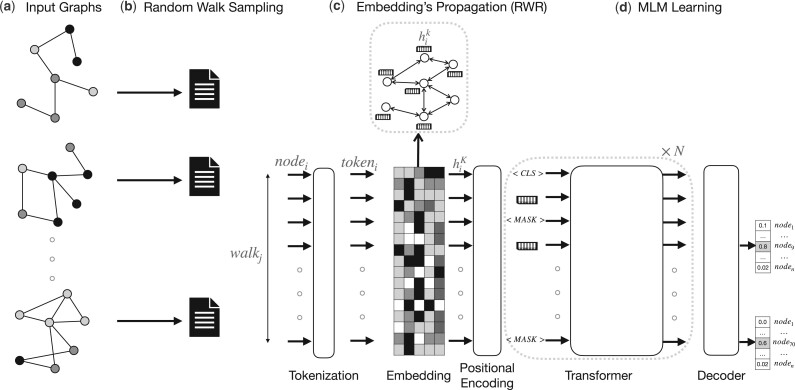
Figure 1.
A sketch of the proposed BERTwalk model. (a and b) Networks are fed into the model and every network is transformed into a collection of node sequences, produced via random walks, and a special symbol [CLS] is added at the beginning of every sequence. (c) The resulting corpus is fed to a transformer model, standard pre-model steps take place in order to numerically represent the data (tokenization and embedding), embedding is propagated every epoch on one of the input networks (iterating over them) before it is fed to the transformer which learns a vector representation for each input that can be used to predict the class of a walk (sequence of nodes) or the identity of masked nodes. (d) The model is trained by masking part of the nodes and training the model to predict them. The end result is an embedding of the nodes, taken from the Embedding layer