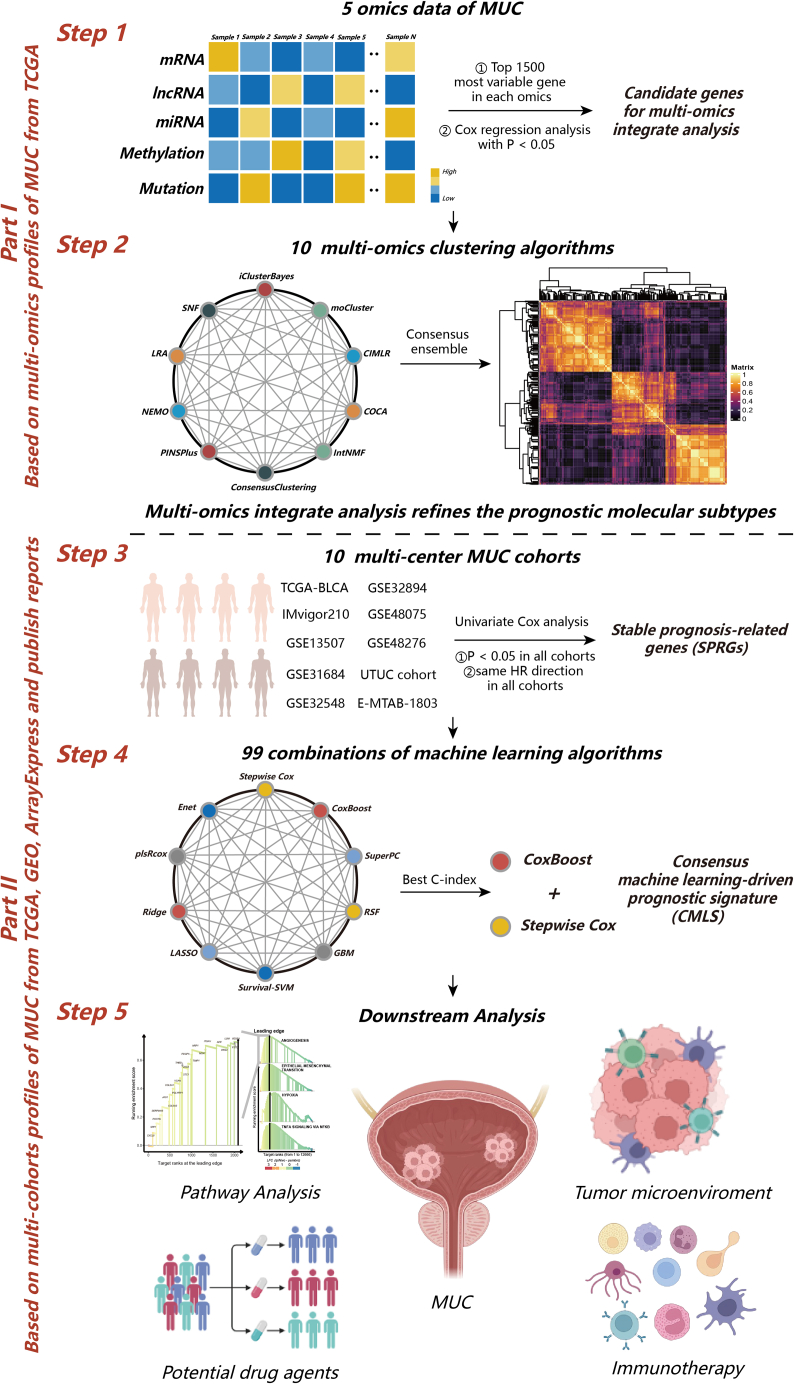
Figure 1.
The computational framework for establishing the CMLS
The top 1,500 genes with the greatest degree of variation were screened from the omics data of each dimension, and then the genes associated with prognosis were selected as clustering candidate genes based on Cox regression analysis. For mutation data, we screened candidate genes based on mutation frequency. Stable MUC prognostic subtypes were obtained by consensus clustering using 10 multiomics clustering algorithms: iClusterBayes, moCluster, Cancer Integration via MultIkernel LeaRning (CIMLR), Integrative Clustering of Multiple Genomic Dataset (IntNMF), ConsensusClustering, Cluster-Of-Cluster-Assignments (COCA), NEighborhood based Multi-Omics clustering (NEMO), PINSPlus, Similarity network fusion (SNF), and Landscape Reconstruction Algorithm (LRA). To further identify stable prognostic genes according to subtypes, all combinations of machine learning algorithms (stepwise Cox, CoxBoost, ridge regression, RSF, GBM, Survival-SVM, LASSO, SuperPC, plsRcox, and Enet) were used to screen out the hub signature with the highest C-index. Finally, CMLS was constructed based on the combination of CoxBoost and stepwise Cox. The relationship between CMLS and prognosis, tumor immune microenvironment, immunotherapy response, and potential therapeutic agents was comprehensively determined.