Abstract
Gastric cancer (GC) is a deadly malignant tumor with a high fatality rate and limited curative options. A growing body of research suggests that network pharmacology can replace traditional methods for determining the precise mechanism of action of medicinal substances in conditions such as cancer. The goal of this study was to clarify the biological mechanism of chelerythrine (CHE) and develop a prediction target for CHE against GC using network pharmacology. First, the genes related to GC were identified from the databases Genecards, Disgenet, Online Mendelian Inheritance in Man, Therapeutic Target Database, and Drugbank, and the targets of CHE were obtained from the SwissTargetPrediction database. Fifty linked targets were identified as anti-GC targets of CHE. Functional enrichment and pathway analyses revealed important biological mechanisms mediated by these targets. The core target PIK3CA of CHE anti-GC was obtained using the protein–protein interaction network, CytoHubba plug-in, and Human Protein Atlas. Molecular docking studies revealed that CHE has a strong affinity for PIK3CA (−10.5 kcal/mol). In addition, we used MTT, colony formation, wound-healing, Transwell®, and flow cytometry experiments to confirm that CHE inhibited the proliferation and migration of GC cells and induced cell cycle arrest and apoptosis. Finally, western blotting results showed that CHE downregulated the expression of the PIK3CA protein and inhibited the activation of the PI3K/AKT signaling pathway. Therefore, we concluded that CHE inhibited GC cell proliferation and migration and induced cell cycle arrest and apoptosis by targeting the PIK3CA protein to inhibit the PI3K/AKT pathway activity.
Keywords: Gastric cancer, Chelerythrine, Network pharmacology, Molecular docking, PIK3CA
Abbreviations
- GC
gastric cancer
- CHE
chelerythrine
- TTD
Therapeutic Target Database
- CC
cellular component
- MF
molecular function
- BP
biological process
- MCC
Maximum Clique Centralit
- DNMNC
Discretized Module Network Centrality
- MNC
Module Network Centrality
- NC
nitrocellulose filter membrane
- PCNA
Proliferating Cell Nuclear Antigen
- EPC
Edge Percolated Component
- HPA
human protein atlas
- SM
mean ± standard error of the mean
- ANOVA
one-way factorial analysis
- GO
gene ontology
- PPI
protein-protein interaction
- FBS
fetal bovine serum
- EMT
epithelial-mesenchymal transition
1. Introduction
Gastric Cancer (GC) remains an important cancer worldwide and is responsible for over one million new cases in 2020 and an estimated 769,000 deaths (equating to one in every 13), globally ranking fifth in incidence and fourth in mortality [1]. Currently available treatment strategies for patients with GC include surgery, chemotherapy, radiotherapy, molecular targeted therapy, and immunotherapy [2]. Nonetheless, GC maintains a high case fatality rate of 75% worldwide and is the main contributor to the global disability-adjusted life-year burden [3]. This is because the onset of GC is insidious. When diagnosed, patients are often in late stages of the disease and lose the best time for treatment. Moreover, the therapeutic efficacy of these agents is limited owing to their associated severe adverse effects, dose-limiting toxicity, drug resistance, and poor selectivity. Therefore, there is an urgent need to develop new therapeutic strategies or new effective drugs to treat GC.
Traditional Chinese medicine monomers have attracted the interest of many researchers because of their low toxicity, high efficiency, multiple targets, and low price [4,5]. Chelerythrine (CHE) is an effective ingredient in the traditional herbal medicine Chelidonium majus L. of the Papaveraceae family, which has a long history of clinical treatment [6,7]. It has been reported to exert antitumor effects in several cancers. CHE showed cytotoxic effects on osteosarcoma cells by inducing apoptosis via activation of the RAF/MEK/ERK pathway [8]. In human renal cell carcinoma, ROS-mediated endoplasmic reticulum stress and STAT3 pathways are the two routes through which CHE promotes apoptosis [9]. CHE also suppressed cell viability and induced apoptosis in A549 and NCI–H1299 NSCLC cells by regulating autophagy [10]. However, the effects and molecular mechanisms of CHE in GC remain largely unknown.
An emerging field called network pharmacology integrates systems biology, pharmacology, computer science, and information networking [11]. Traditional bioinformatics has been used to help uncover the underlying processes between medicinal molecules and potential targets [12]. Network pharmacology, used in targeted therapy, uses network component analysis to characterize intricate interactions between biological systems to identify synergistic effects in cancer treatment [13]. Molecular docking is a method used to design drugs by simulating interactions between receptors and drugs. In recent years, the application of molecular docking technology to explain related mechanisms has become a trend in new drug research and development [13].
In this study, CHE was found to have a significant inhibitory effect on HGC-27 and MGC-803 cells in the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. Network pharmacology was used to analyze the biological processes and pathways. Molecular docking was used to verify the targets obtained from previous studies. Simultaneously, the targets with the closest correlation with CHE were screened using the dock-binding free energies. Finally, we used colony formation, apoptosis, wound-healing, Transwell®, and western blotting assays to confirm the network pharmacology and molecular docking results. The flowchart of the study is shown in Fig. 1.
Fig. 1.
Flowchart of this study.
2. Materials and methods
2.1. Data collection and preparation
Briefly, the screening of targets against GC hits and the relative candidate targets of CHE were identified using databases. First, the Swiss Target Prediction database (http://swisstargetprediction.ch/) was used to predict the putative targets of CHE (targets with a probability of 0 were deleted). A drug-target diagram was constructed using Cytoscape 3.9.1 [14]. Second, the therapeutic targets connected with GC were collected from the Genecards database (targets with Relevance score <1 were excluded) (https://www.genecards.org/), Disgenet database (targets with DSI_g less than 0.1 were discarded) (http://www.disgenet.org/), Drugbank database (https://go.drugbank.com/), Therapeutic Target Database (TTD) (http://db.idrblab.net/ttd/), and KEGG database (https://www.genome.jp/kegg/disease/) using the keyword search 'gastric cancer.' Targets that appeared more than twice were considered GC hits. The intersection results are shown in the VEEN diagram. Third, the overlapping GC hits with those of the CHE targets were considered as anti-GC CHE targets and were considered for further analysis. Finally, VEEN plots created using R (version 4.0.3) were used to visualize the intersection targets.
2.2. Gene ontology (GO) analysis and pathway enrichment on the identifiable targets
Gene Ontology and functional pathway enrichment of the obtained targets were performed using R. The screening standard was set as p = 0.05 and q = 0.05’ for the functional enrichment analysis results. The top 10 results of cellular component (CC), molecular function (MF), and biological process (BP) by importance in GO analysis were analyzed using R to create bubble charts. The top 30 KEGG pathway enrichments were used to draw a bubble diagram for visual display.
2.3. Construction of a protein–protein interaction (PPI) network and identification of core target
The PPI network was constructed using STRING v11.5 (https://cn.string-db.org) by importing the CHE anti-GC-related hits from Method 2.1. Homo sapiens was selected as the background species for the genes. We set the minimum necessary interaction score to be the 'highest confidence (0.9)' and hidden disconnected nodes in the network to obtain more convincing results. STRING networks were imported into Cytoscape (version 3.9.1). The top five were obtained according to the six algorithms of Maximum Clique Centralit (MCC), Discretized Module Network Centrality (DNMNC), Module Network Centrality (MNC), Degree Centrality (Degree), Edge Percolated Component (EPC), and BottleNECK in CytoHubba, and the intersection is depicted by an UpSet diagram. Targets that appeared six times were considered core targets.
2.4. Evaluation of target protein expression level in GC tissues and its effect on overall survival prognosis of GC patients
The Human Protein Atlas (HPA) database (https://www.proteinatlas.org/) was used to examine the expression levels of the target proteins. HPA uses a mix of antibody-based proteomics and transcriptomics to map the human proteome to a single-cell level. More than 13 million distinct immunohistochemistry images are included in a comprehensive, freely accessible database, which is a great resource for exploring and researching potential therapeutic targets and disease biomarkers [15]. Immunohistochemistry was used to compare the expression levels of the target proteins between GC and normal gastric cells. The five-year survival rates of patients with high and low expression of the target proteins were analyzed.
2.5. Analysis via molecular docking
Utilizing the AutoDock Vina 1.1.2 molecular docking software, potential targets suggested from the network analysis were evaluated. The sdf format of CHE was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/), and the energy of the structure was minimized using Chembio3D Ultra 14.0. In addition, the 3D structures of possible targets were obtained from the Protein DataBank database (https://www.rcsb.org/). All water molecules and binding substances were removed using PyMOL. AutoDock Tools 1.5.6 software was used to add all the hydrogens, calculate Gasteiger charges for the structure, and save them as receptors in the PDBQT file format. Finally, AutoDock Vina 1.1.2 was used for docking, and PyMOL software was used for visualization. When the binding energy was < −7.0 kcal/mol, we assumed a strong binding affinity between the targets and CHE.
2.6. Cell culture
The human undifferentiated GC cell line HGC-27 (Aoyin Biotechnology Co., Ltd, Shanghai, China) and the gastric adenocarcinoma cell line MGC-803 (donated by Dr. Zhu Mingwei, Department of Abdominal Ultrasound, Harbin Medical University) were cultured in Roswell Park Memorial Institute 1640 medium and DMEM containing 10% heat-inactivated fetal bovine serum (FBS), and 1% double antibody (penicillin and streptomycin), respectively. Cells were maintained at 37 ᵒC in a humidified environment composed of 95% air and 5% CO2. CHE (≥95% purity) was purchased from Aladdin (Shanghai, China).
2.7. Pharmacological study by in vitro cytotoxicity assessment
HGC-27 and MGC-803 cells were cultivated separately in 96-well plates at 4 × 103 cells per well. The cells in exponential growth phase were treated with different doses of CHE (0, 2, 4, 6, 8, 10, 12, 16, 18, and 20 μmol/L) and incubated during two distinct periods of 24 and 48 h. After incubation for 4 h in the dark, the MTT assay was used to determine the viability of the cells. The formation of formazan crystals was measured spectrophotometrically after solubilization in DMSO at 490 nm. The number of viable cells was determined by comparison with the proportion of the untreated cells. Three replicates were used in this experiment.
2.8. Plate colony formation assay
HGC-27 and MGC-803 cells (500 cells/well) were seeded in 6 cm dishes. After incubation for 24 h, the cells were treated with CHE (0 and 10 μmol/L) for 1 h, after which the normal complete culture medium was changed. After culturing for 10 days, the cell colonies were first fixed with methanol and then stained with 0.5% crystal violet for 20 min at room temperature (approximately 25 ᵒC), counted, and quantified using Image J (National Institutes of Health, Bethesda, MD, USA) [16]. Three replicates were used in this experiment.
2.9. Wound-healing assays
HGC-27 and MGC-803 cells were grown to 100% confluence for wound-healing assays. Confluent monolayer GC cells were scraped linearly with the help of a 10 μL pipette tip to create an artificial scratch. Subsequently, 1 × PBS was added to the wells to wash them several times and remove floating cells. These cells were then exposed to medium supplemented with CHE (0 and 2 μmol/L) for 24 h. Images were obtained at 0, 12, and 24 h using a microscope to analyze the movement of cells to close the wound. ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to measure and analyze the migratory distance.
2.10. Transwell® assays
The Transwell® assay was used to evaluate the effect of CHE on the migration and invasion of GC cells. In the migration experiment, GC cells with the concentration set were seeded in the upper chamber of the Transwell® chamber and incubated with 5% FBS medium, and medium containing 10% FBS was injected into the lower chamber. After incubation for 24 h, cells were removed from the culture medium. Subsequently, cells attached to the lower surface of the chamber were fixed with methanol. Finally, the migrated GC cells were stained with 0.5% crystal violet for 20 min. Images were collected and analyzed using the previously mentioned microscope. The experiment was independently repeated three times.
2.11. Cell cycle assay
HGC-27 and MGC-803 cells were plated at a density of 3 × 105 cells per well in 6-well culture plates and allowed to develop until 90% confluency. The cells were then exposed to various concentrations of CHE (0 and 10 μmol/L) and incubated for 24 h. Finally, cells were harvested and incubated with 1 mL DNA staining solution and 10 μL permeabilization solution (MULTISCIENCES, Hangzhou, China). Three replicates were used in this experiment.
2.12. Cell apoptosis assay
GC cells were treated with CHE (0, 5, 10, and 20 μmol/L) for 24 h, and adherent and floating cells were washed with PBS and harvested with trypsin. The washed cells were then collected by centrifugation and resuspended in staining buffer containing 5 μL annexin V and 10 μL PI (200 μg/mL) (4A Biotech, Beijing, China). Following incubation of the cells at room temperature for 5 min in the dark, the proportion of cells at different stages of apoptosis was quantified using flow cytometry. This experiment was performed in triplicate.
2.13. Detection of target and phenotype-related proteins
Western blot analysis was performed to determine the in vitro protein expression of critical targets. HGC-27 and MGC-803 cells were seeded on 6.0 cm plates at a density of 3 × 105 cells per well and then exposed to CHE at the stated concentrations (0, 5, 10, and 20 μmol/L) for 24 h at 37 °C. Radioimmunoprecipitation assay buffer (BCA, Beyotime Biotechnology, Shanghai, China) containing protease and phosphatase inhibitors was used to extract total protein from each group after the cells were collected. The amount of protein in each group was calculated using the Pierce™ BCA assay (Beyotime Biotechnology, Shanghai, China). Based on their molecular weights, the protein samples were separated by 10–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to a nitrocellulose filter membrane (NC). A 5% solution of bovine serum albumin was used to block the membrane for approximately 1 h at room temperature. After blocking, each target protein was incubated with the following primary antibodies: Proliferating Cell Nuclear Antigen, PCNA) (diluted 1:750, rabbit pAb, Cat: WL05017, WanLei Biological Technology Co., Ltd., Shenyang, China), BAX (diluted 1:1000, rabbit pAb, cat No. 60267-1-Ig, Proteintech Technology Co., Ltd., Wuhan, China), BCL2 (diluted 1:750, rabbit pAb, Cat: WL05017, WanLei Biological Technology Co., Ltd., Shenyang, China), cleaved-caspased 9 (diluted 1:1000, rabbit pAb, cat No. 20750S, Cell Signaling Technology Co., Ltd., Boston, MA, USA), vimentin (diluted of 1:2000, rabbit pAb, cat No. 10366-1-AP, Proteintech Technology Co., Ltd., Wuhan, China), N-cadherin (diluted 1:1000, rabbit pAb, cat No. 22018-1-AP, Proteintech Technology Co., Ltd., Wuhan, China), AKT (diluted 1:1000, rabbit pAb, cat No. 9372S, Cell Signaling Technology Co., Ltd., Boston, MA, USA), p-AKT (diluted 1:1000, rabbit pAb, cat No. 4060S, Cell Signaling Technology Co., Ltd., Boston, MA, USA), PIK3CA (diluted 1:500, rabbit pAb, Cat: WL05017, WanLei Biological Technology Co., Ltd., Shenyang, China), and β-actin (diluted 1:1000, mouse pAb, cat No. TA-09, Origene Technologies, Inc., Beijing, China) overnight at 4 °C. Membranes that had been incubated were routinely washed with PBS-T solution at least five times, with each wash lasting 15 min. After thorough cleaning, the membranes were incubated with secondary antibody (LI-COR Biosciences, Lincoln, NE, USA) for 1 h at room temperature. An Odyssey FC imaging system (LI-COR Biosciences, Lincoln, NE, USA) was used to visualize and scan the blots. Protein expression was examined using ImageJ 6.0 (National Institutes of Health, Bethesda, MD, USA). β-actin was used as a control to standardize the relative density of the protein bands.
2.14. Statistical analysis
The experimental data were statistically analyzed using R or GraphPad Prism (version 8.0, GraphPad, Inc., San Diego, CA, USA). The results are presented as the mean ± standard error of the mean (SEM) of triplicate samples. Student's t-test was used after one-way factorial analysis (ANOVA) to determine significant differences between groups. *p value of <0.05 and **p value of <0.01, were considered statistically significant.
3. Results
3.1. Target identification of anti-GC targets of CHE
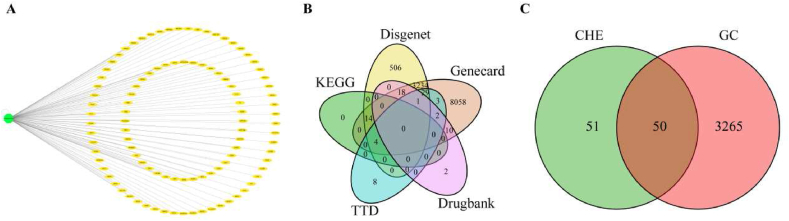
We obtained 101 potential targets for CHE from the SwissTargetPrediction database and presented them using a drug-target network diagram (Fig. 2A, Supplementary Table 1). From other databases, 11373, 3806, 33, 47, and 18 targets were screened from Genecards, Disgenet, Drugbank, TTD, and KEGG, respectively (Supplementary Table 2). The intersectional targets are shown in the VEEN diagram (Fig. 2B, Supplementary Table 3). Subsequently, 3315 targets that met our requirements (repeated more than twice) were considered GC-related therapeutic targets. Repeated targets of CHE and GC-related therapeutic targets are shown in the VEEN diagram (Fig. 2C, Supplementary Table 4). Finally, 50 anti-GC targets of CHE were identified.
Fig. 2.
Related targets of CHE anti-GC. (A) Network diagram of potential CHE targets. (B) Veen diagram depicting the intersection of GC-related targets in the five databases. (C) Venn diagram showing the intersection of GC-related therapeutic targets and potential CHE targets.
GC, gastric cancer; CHE, chelerythrine.
3.2. Targets associated with the anti-GC role of CHE were analyzed for function and KEGG pathway enrichment
The R software was used to assess the biological processes and enhanced signaling pathways of all 50 anti-GC targets of CHE (Fig. 3A, Supplementary Table 5). The results showed that CHE was involved in protein binding, ATP binding, serine/threonine kinase activity, protein kinase activity, and other MFs through cell sites such as cytosol, nucleus, cytoplasm, plasma membrane, mitochondria, and other CC, to affect protein phosphorylation, negative regulation of apoptosis, signal transduction, drug response, apoptosis, and positive and negative regulation of RNA polymerase II promoter transcription. In addition, in KEGG pathway analysis, 128 KEGG pathways related to all core targets were identified (p adjust <0.05) (Fig. 3B, Supplementary Table 6). The top ten included neutrophil extracellular trap formation, viral carcinogenesis, microRNAs in cancer, the PI3K-Akt signaling pathway, the thyroid hormone signaling pathway, and cellular senescence. We analyzed the first ten pathways and found that the cancer pathway was more prevalent, with enriched 30 genes, and the P13K-Akt signaling pathway, with 12 genes. The PI3K/AKT pathway is a classic tumor pathway, indicating that CHE plays an anti-GC role.
Fig. 3.
(A) Enrichment of gene ontology in CHE anti-GC prediction target biological processes, cell composition, and molecular function. (B) Pathway enrichment analysis of CHE-predicted targets ranked among the top 30.
GC, gastric cancer; CHE, chelerythrine.
3.3. PPI network analysis and determination of CHE anti-GC core targets
A PPI network (Fig. 4A) was constructed with highly reliable (>0.9) interaction goals, including 50 nodes and 47 edges. The average node degree, local clustering coefficient, and PPI enrichment p values of the network were 1.88, 0.349, and 2.47e-07, respectively. The network was then imported into Cytoscape 3.9.1 (Fig. 4B–G, Supplementary Table 7). The top five targets were screened using the MCC, DMNC, MNC, DEGREE, EPC, and BottleNECK algorithms in the CytoHubba plug-in (Supplementary Table 8) [17]. We used the up-set graph (Fig. 4H, Supplementary Table 9) to analyze the top five targets screened by the different algorithms, and the core targets appeared in the top five targets of the six algorithms. PIK3CA and PDPK1 are considered key targets for the CHE treatment of GC.
Fig. 4.
Construction of the PPI network of CHE treatment GC-related targets and determination of core targets. (A) The PPI network of GC-related targets constructed by CHE treatment. (B–G) The top five targets were screened using the MCC, DMNC, MNC, DEGREE, EPC, and BottleNECK algorithms in the CytoHubba plug-in. The depth of the color represents the importance of the target in each algorithm. (H) The intersection of the top five targets in the six algorithms was visualized using up-set plots. The blue column in the lower left corner represents the name of the algorithm, the blue column in the upper right corner represents the number of repetitions of the target, and the origin in the lower right corner represents the specific information of the repetition of the specific target.
GC, gastric cancer; CHE, chelerythrine; PPI, protein-protein interaction.. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Verification of the HPA database
We used the HPA database to analyze the expression of PIK3CA and PDPK1 in GC and normal gastric tissues (Fig. 5A, C). First, we found that the expression of PIK3CA and PDPK1 proteins was higher in gastric cancer tissues than in normal gastric tissues. Second, we found that low expression of PI3KCA (with a cutoff value of 3.85) was associated with a higher survival rate than high expression of PIK3CA (p = 0.012) (Fig. 5B). The expression of PDPK1 (with a cut-off value of 2.6) seems to play different roles at different stages of the disease. Patients with high expression of PDPK1 protein have a higher survival rate when the survival time of patients is less than five years, and patients with PDPK1 protein have a lower survival rate when the survival time is over five years, which means that the function of PDPK1 may be diverse (Fig. 5D). Combined with the screening of core targets and verification using the HPA database, we believe that PIK3CA is the most important target of CHE in GC. This was consistent with our KEGG pathway enrichment analysis results, indicating that the PI3K/AKT pathway plays an important role in the treatment of GC with CHE.
Fig. 5.
HPA data validation of core targets. (A, C) Immunohistochemical staining results of PIK3CA protein expression in normal gastric and cancer tissues (scale bar represents 200 μm). (B, D) Expression of PIK3CA and PDPK1 and the prognosis of patients with GC.
3.5. Verification of docking simulation
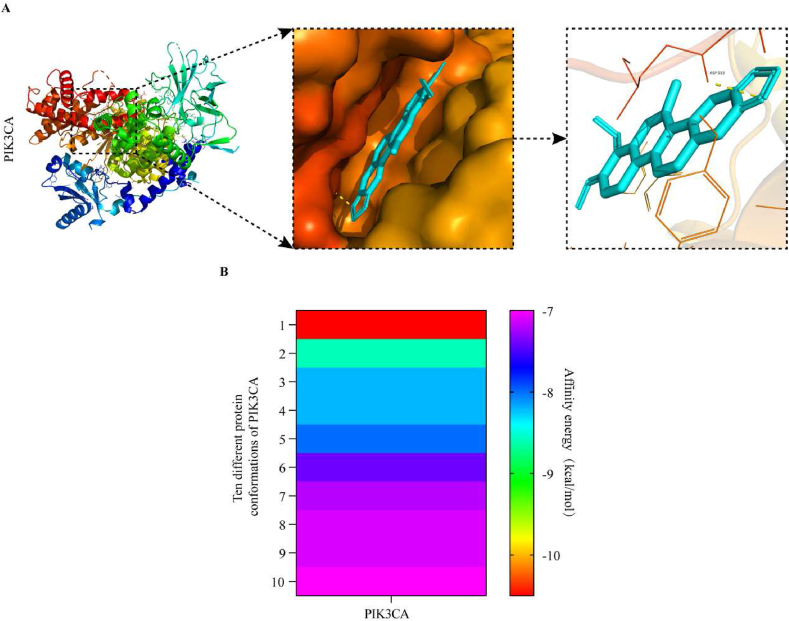
To evaluate the regulation of CHE on the core target, PIK3CA, we used molecular docking to identify the binding ability between CHE and PIK3CA (PBD ID:6PSY). As shown in Fig. 6A (and Supplementary Table 10), CHE has strong binding activity with PIK3CA (affinity energy = −10.5 kcal/mol). Fig. 6B shows the energy heat map of CHE and PIK3CA binding to the top ten in different conformations. The optimal binding energy was −10.5 kcal/mol, and the weakest binding energy was −7.0 kcal/mol, indicating that CHE and PIK3CA have strong coupling.
Fig. 6.
Molecular docking of PIK3CA with CHE. (A) Binding poses of CHE complexed with PIK3CA, optimal affinity = −10.5 kcal/mol. (B) Top ten binding energy heat maps of CHE with different conformations of PIK3CA. The affinities range from −10.5 kcal/mol to −7.0 kcal/mol.
GC, gastric cancer; CHE, chelerythrine.
3.6. In vitro verification of the pharmacological toxicity of CHE on GC cell lines
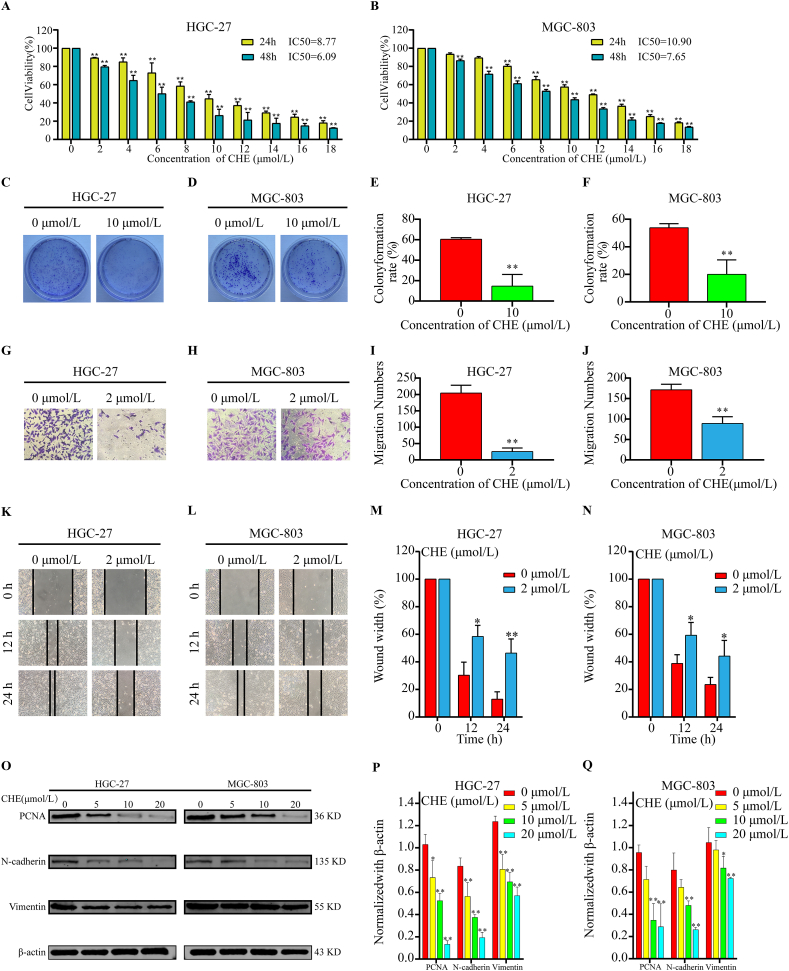
To further validate the bioinformatic results, at two-time intervals (24 and 48 h), the cytotoxicity of various doses of CHE (0–18 μmol/L) on HGC–27 and MGC–803 cells was assessed using the MTT assay. The viability of GC cells was considerably decreased by CHE in a dose- and time-dependent manner (Fig. 7A and B). The two GC cells' respective IC50 values for CHE were determined to be 8.77 μmol/L in HGC-27 cells and 10.90 μmol/L in MGC-803 cells at 24 h. Additionally, HGC-27 cells were more sensitive to the cytotoxic effects of CHE at lower concentrations than MGC-803 cells. A colony is a collection of cells made up of a single cell's progeny that has multiplied in vitro for more than six generations. Colony formation rate reflects stemness and cell proliferation. Fig. 7 (C–F) demonstrates that CHE-treated GC cells generated fewer colonies than untreated cells, indicating that CHE treatment slowed the growth of GC. This conclusion was further supported by the fact that CHE treatment reduced the production of the proliferation marker protein, PCNA (Fig. 7O and Fig. S1A).
Fig. 7.
CHE decreases the activity, proliferation, and migration of GC cells. (A, B) GC cell survival after exposure to CHE for 24 and 48 h. (C, D) The capacity of GC cells treated with various doses (0 and 10 μmol/L) of CHE to produce clones was assessed. (E, F) Equivalent colony development rates determined by statistical analysis of three different experiments. (G, H) The mobility of GC cells treated with CHE in three-dimensional space was assessed. From the statistical findings of the three separate trials, we can derive the mobility measurements in (I) and (J). (K, L) Determination of healing rate of GC cells treated with CHE. (M, N) Statistical results from three separate studies were used to calculate the relevant healing rate. (O) Protein expression of PCNA, N-cadherin, and vimentin in GC cells after CHE treatment. The statistical results of the relative protein expression levels from three separate studies are shown in (P) and (Q). *p < 0.05, * *p < 0.01 in contrast to the control group.
GC, gastric cancer; CHE, chelerythrine.
Chemotherapeutic medications that impair cell viability typically affect cell motility. Using a wound-healing test, the motility of GC cells was evaluated at low CHE concentrations (0–2 μmol/L) without endangering their survival. Fig. (7G–J) demonstrates that, whereas scratches in the control group steadily healed over the course of 24 h, the procedure was considerably delayed after the administration of CHE. Fewer cells consistently entered the bottom chamber through the hole when CHE levels increased in the CHE intervention group according to Transwell® testing (Fig. 7K-N). As epithelial-mesenchymal transition (EMT) has been demonstrated to be one of the crucial mechanisms of cancer cell invasion and migration, we used western blotting to evaluate the expression of EMT-related markers. N-cadherin and vimentin expression in GC cells exposed to CHE was decreased, as shown in Fig. 7O (and Fig. S1A). Additionally, the changes in protein expression were dose-dependent. These findings imply that CHE prevents GC cell migration and slows the EMT.
3.7. CHE induced G0/G1 arrest and apoptosis in GC cells
The results of the cell cycle assay showed that CHE blocked the cell cycle of GC cells in the G0/G1 phase (Fig. 8A-D). As cell proliferation depends on the cell cycle, we concluded that CHE could hinder the proliferation of GC cells by blocking the cells in the G0/G1 phase of the cell cycle. This is consistent with the results of the GO enrichment analysis.
Fig. 8.
Apoptosis and cell cycle changes in GC cells after CHE treatment. (A, B) Cell cycle assays of HGC and MGC-803 cells treated with CHE (10 μmol/L). (C, D) The corresponding G0/G1 phase cell ratios from the statistical results of three independent experiments. (E) Apoptosis tests on GC cells that had been treated with CHE and (F, G) the matching apoptosis rate derived from the statistical findings of three separate studies. (H) PCNA protein expression in GC cells following CHE treatment and (I, J) the related statistical findings of the relative protein expression values from three separate studies. *p < 0.05, **p < 0.01.
GC, gastric cancer; CHE, chelerythrine; PCNA, Proliferating Cell Nuclear Antigen.
In the MTT assay, GC cells incubated with CHE showed obvious morphological changes and were mainly contracted and floated. These observations suggested that CHE has potential apoptotic activity in GC cells. Using Annexin V-FITC/PI staining and flow cytometry analysis, we detected and demonstrated that CHE is a significant apoptosis-inducing factor in GC cells (Fig. 8E). The apoptosis rates of HGC-27 and MGC-803 cells treated with 20 μmol/L CHE were 10.8 times and 5.06 times higher, respectively, than those of the corresponding control groups (Fig. 8F–G). In addition, the expression of Bcl-2 (an anti-apoptotic marker) in CHE-treated GC cells decreased, while the expression of Bax and cleaved-caspased 9 (a pro-apoptotic marker) increased (Fig. 8H–J and Fig. S1B), indicating that CHE-treated GC cells undergo apoptosis through intrinsic apoptotic pathways.
3.8. Verification of protein levels of core targets and downstream effect targets
Western blotting was used to detect the key factors of the PI3K/AKT pathway in GC cells, including PIK3CA and AKT, and its corresponding phosphorylated form, p-AKT. PIK3CA is located on chromosome 3 and contains 20 exons that encode the p110 catalytic subunit of class I phosphatidylinositol-3-kinase, namely PI3Kp110α. The results showed that CHE significantly reduced the expression of p-AKT and PIK3CA in a dose-dependent manner (Fig. 9A and Fig. S1C). However, no significant difference was observed in the total AKT expression.
Fig. 9.
Expression of PIK3CA and pAKT/AKT after CHE treatment. (A) The protein expression of p-AKT, AKT, and PIK3CA following CHE treatment and (B, C) the corresponding statistical findings of the relative protein expression values obtained from three separate trials. *p < 0.05, **p < 0.01.
CHE, chelerythrine.
4. Discussion
With a mortality rate in the later stages, GC is the third most common cause of cancer cell death worldwide. The initial diagnosis, course, and therapy of this disease are often poor [2]. Although chemotherapy and immunotherapy are effective against the illness, recent trends have shifted toward the utilization of natural therapies in expanding studies [18,19]. In this study, a bioinformatics inquiry from web-based databases using a network pharmacological method was used to investigate the therapeutic mechanism of the natural alkaloid compound CHE as a therapy for GC. To the best of our knowledge, this is the first study to combine network pharmacology, molecular docking simulations, and experimental validation to elucidate the pharmacological mechanisms of CHE against GC.
Database screening was primarily used to identify the treatment targets of CHE and GC. Consequently, 50 possible targets of CHE in GC were identified. The selected targets were examined for functional enrichment using GO analysis. Protein binding, ATP binding, serine/threonine kinase activity, and the molecular role of protein kinase activity were the most often linked to enriched target categories. Furthermore, according to KEGG pathway enrichment analysis, the PI3K/AKT signaling pathway contained the majority of treatment-related targets.
Numerous studies have demonstrated a strong association between PIK3CA and tumor growth. A phase II clinical study published in The Journal of Clinical Investigation showed that PIK3CA inhibitors could benefit patients with head and neck tumors with atypical PIK3CA mutations [20]. Studies have shown that the combination of alpelisib-fulvestrant can prolong progression-free survival in patients with advanced breast cancer with PIK3CA3 mutation, HER2 positive, HER2 negative, and endocrine therapy [21,22]. Wu et al. showed that PI3K/AKT/mTOR activation induced by PIK3CA upregulation promotes GC progression and chemoresistance [23]. A meta-analysis showed that high PIK3CA expression is closely related to poor prognosis in patients with gastric cancer [24]. Therefore, targeting PIK3CA is an effective strategy for increasing the survival time of patients with GC.
To properly screen the primary CHE targets for treating GC, we used the top five targets obtained by the six algorithms in the CytoHubba plug-in and repeated the targets that appeared six times as the key targets of CHE in the treatment of GC. Finally, we identified PIK3 CA and PDPK1. Interestingly, this was consistent with the results of the KEGG enrichment analysis. Subsequently, the clinical significance of the target was analyzed using the HPA database. We discovered a strong correlation between low PIK3CA protein expression and the five-year survival rates of patients with GC. Thus, PDPK1 may play different roles at different stages of the disease. Functional analysis of PIK3CA and PDPK1 using Genecards revealed that PIK3CA can function as a catalytic subunit of PI3K, while PDPK1, and p85, function as accessory subunits. Therefore, we believe that PIK3CA is a core target of CHE in GC. In summary, we concluded that CHE may control PI3K/Akt/mTOR signaling pathway activation by interacting with PIK3CA.
Many studies have suggested that the PI3K/AKT signaling pathway is associated with tumor cell proliferation, migration, and apoptosis [[25], [26], [27]]. We found that CHE inhibited the activity and proliferation of HGC-27 and MGC-803 cells using MTT and colony formation assays. Moreover, Western blot analysis revealed lower levels of PCNA, a protein associated with cell proliferation. To avoid the effect of tumor cell proliferation on migration experiments, we used western blotting to detect the protein expression of interstitial markers N-cadherin and vimentin. The results showed that the expression of interstitial markers gradually decreased with increasing CHE concentrations. Flow cytometry was used to analyze cell cycle changes and apoptosis rates after CHE treatment of GC cells. These findings demonstrated that CHE caused G0/G1 arrest in GC cells and, in a concentration-dependent manner, enhanced GC cell apoptosis. After CHE treatment of GC cells, we found that the expression of the anti-apoptotic protein Bcl2 decreased, while the expression of pro-apoptotic proteins Bax and cleaved caspase 9 increased. In summary, CHE inhibited GC cell growth by inhibiting GC cell activity and proliferation and inducing G0/G1 phase cell arrest and apoptosis.
Finally, to explore the regulation of the PIK3CA protein by CHE, we first analyzed the affinity between CHE and PIK3CA by molecular docking. The results showed that the binding energy between CHE and PIK3CA was −10.5 kcal/mol, and CHE formed hydrogen bonds with residues of PIK3CA, namely Asp933. Studies have shown that when the binding energy is less than −7.0 kcal/mol, there is strong coupling between small molecules and proteins. This indicated that PIK3CA has a strong affinity for CHE. Our Western blot results also demonstrated this. In addition, we detected the expression levels of the PIK3CA downstream target, AKT, and its phosphorylated form, pAKT. With an increase in CHE, the expression level of pAKT gradually decreased, but the change in AKT was not statistically significant. In summary, CHE inhibited GC cell viability and proliferation, and induced G0/G1 phase arrest and apoptosis by targeting PIK3CA to downregulate the activity of the PI3K/AKT signaling pathway, ultimately inhibiting GC cell growth.
The clinical transformation of drugs and verification of their in vivo efficacy are necessary steps. Unfortunately, owing to the novel coronavirus epidemic, we were unable to conduct in vivo experiments. However, previous studies have shown that CHE still has a good therapeutic effect in in vivo studies of glioma, non-small cell lung cancer, and Dalton's lymphoma [28,29], suggesting that CHE may also have good pharmacological activity in in vivo studies of GC. In addition, chemotherapy tolerance is one of the most important factors leading to poor prognosis in patients with gastric cancer. Many studies have found that abnormal activation of the PI3K/AKT signaling pathway is involved in chemotherapy tolerance in GC. Thus, CHE combined with platinum or fluorouracil may have better anti-GC activity. In summary, CHE has a great clinical value as an anti-GC drug.
5. Conclusions
This study systematically elucidated the mechanism of action of CHE against GC by network pharmacology, molecular docking, and experimental verification. CHE causes cell cycle arrest and death and reduces the growth and migration of GC cells. This mechanism may be related to the inhibition of PI3K/AKT signaling pathway activation, which is mediated by targeting PIK3CA. In addition, given the important role of PIK3CA in the progression of gastric cancer, CHE has strong drug development potential and can be used as a therapeutic drug for patients with GC.
Author contribution statement
Kang Kai, M.D; Jiang Hanbing, M.D: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Cheng Binglin, Ph.D.: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Zhang shujun: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data will be made available on request.
Additional information
No additional information is available for this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The author thanks Ms. Mingwei zhu for her helpful suggestions on experimental technology. This research was completed in the Hepatosplenic Surgery Laboratory of the First Affiliated Hospital of Harbin Medical University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17393.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Smyth E.C., Nilsson M., Grabsch H.I., van Grieken N.C., Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 3.Thrift A.P., El-Serag H.B. Burden of gastric cancer. Clin. Gastroenterol. Hepatol. 2020;18:534–542. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuli H.S., Mittal S., Aggarwal D., Sharma .A.K. Path of Silibinin from diet to medicine: a dietary polyphenolic flavonoid having potential anti-cancer therapeutic significance. Semin. Cancer Biol. 2021;73:196–218. doi: 10.1016/j.semcancer.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Salehi B., Varoni E.M., Sharifi-Rad M., Rajabi S., Zucca P., Iriti M., Sharifi-Rad J. Epithelial-mesenchymal transition as a target for botanicals in cancer metastasis. Phytomedicine. 2019;55:125–136. doi: 10.1016/j.phymed.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Zielinska S., Jezierska-Domaradzka A., Wojciak-Kosior M., Sowa I., Junka A., Matkowski A.M. Greater celandine's ups and downs-21 centuries of medicinal uses of chelidonium majus from the viewpoint of today's pharmacology. Front. Pharmacol. 2018;9:299. doi: 10.3389/fphar.2018.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilca M., Gaman L., Panait E., Stoian I., Atanasiu V. Chelidonium majus--an integrative review: traditional knowledge versus modern findings. Forsch Komplementmed. 2010;17:241–248. doi: 10.1159/000321397. [DOI] [PubMed] [Google Scholar]
- 8.Yang R., Piperdi S., Gorlick R. Activation of the RAF/mitogen-activated protein/extracellular signal-regulated kinase kinase/extracellular signal-regulated kinase pathway mediates apoptosis induced by chelerythrine in osteosarcoma. Clin. Cancer Res. 2008;14:6396–6404. doi: 10.1158/1078-0432.CCR-07-5113. [DOI] [PubMed] [Google Scholar]
- 9.He H., Zhuo R., Dai J., Wang X., Huang X., Wang H., Xu D. Chelerythrine induces apoptosis via ROS-mediated endoplasmic reticulum stress and STAT3 pathways in human renal cell carcinoma. J. Cell Mol. Med. 2020;24:50–60. doi: 10.1111/jcmm.14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Z.H., Cao W.X., Wang Z.Y., Lu J.H., Liu B., Chen X., Lu J.J. Induction of reactive oxygen species-stimulated distinctive autophagy by chelerythrine in non-small cell lung cancer cells. Redox Biol. 2017;12:367–376. doi: 10.1016/j.redox.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopkins A.L. Network pharmacology. Nat. Biotechnol. 2007;25:1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 12.Zeng L., Yang K., Ge J. Uncovering the pharmacological mechanism of astragalus salvia compound on Pregnancy-Induced hypertension syndrome by a network pharmacology approach. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-17139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakle N.S., More S.A., Mokale S.N. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: an updated prototype in drug discovery. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-74251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shannon P., Markiel A., Ozier O., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindskog C. The potential clinical impact of the tissue-based map of the human proteome. Expert Rev. Proteomics. 2015;12:213–215. doi: 10.1586/14789450.2015.1040771. [DOI] [PubMed] [Google Scholar]
- 16.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Lou Y., Wang J., Yu C., Shen W. Research status and molecular mechanism of the traditional Chinese medicine and antitumor therapy combined strategy based on tumor microenvironment. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.609705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W., Li B., Xu M., Yang T., Hao X. Traditional Chinese medicine for precancerous lesions of gastric cancer: a review. Biomed. Pharmacother. 2022;146 doi: 10.1016/j.biopha.2021.112542. [DOI] [PubMed] [Google Scholar]
- 20.Jin N., Keam B., Cho J., Grandis .J.R. Therapeutic implications of activating noncanonical PIK3CA mutations in head and neck squamous cell carcinoma. J. Clin. Invest. 2021;131 doi: 10.1172/JCI150335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agahozo M.C., Sieuwerts A.M., Doebar S.C., van Deurzen .C. PIK3CA mutations in ductal carcinoma in situ and adjacent invasive breast cancer. Endocr. Relat. Cancer. 2019;26:471–482. doi: 10.1530/ERC-19-0019. [DOI] [PubMed] [Google Scholar]
- 22.Andre F., Ciruelos E., Rubovszky G., Juric D. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 23.Wu Q., Ma J., Wei J., Meng W., Wang Y., Shi M. FOXD1-AS1 regulates FOXD1 translation and promotes gastric cancer progression and chemoresistance by activating the PI3K/AKT/mTOR pathway. Mol. Oncol. 2021;15:299–316. doi: 10.1002/1878-0261.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Chen S., Li H., Huo .Z. Association between PIK3CA alteration and prognosis of gastric cancer patients: a meta-analysis. Oncotarget. 2018;9:7651–7659. doi: 10.18632/oncotarget.23871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S., Chen M., Huang J., Lam .K.O. ORAI2 promotes gastric cancer tumorigenicity and metastasis through PI3K/Akt signaling and MAPK-Dependent focal adhesion disassembly. Cancer Res. 2021;81:986–1000. doi: 10.1158/0008-5472.CAN-20-0049. [DOI] [PubMed] [Google Scholar]
- 26.Ao R., Guan L., Wang Y., Wang J.N. Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell proliferation, migration, and invasion while promoting apoptosis through the PI3k-Akt signaling pathway. J. Cell. Biochem. 2018;119:4420–4434. doi: 10.1002/jcb.26524. [DOI] [PubMed] [Google Scholar]
- 27.Yan H.Z., Wang H.F., Yin Y., He .B.S. GHR is involved in gastric cell growth and apoptosis via PI3K/AKT signalling. J. Cell Mol. Med. 2021;25:2450–2458. doi: 10.1111/jcmm.16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Song Y., Zhang N., Li N., Liu C., Wang B. Using liposomes to alleviate the toxicity of chelerythrine, a natural PKC inhibitor, in treating Non-Small cell lung cancer. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.658543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu M., Niu J., Jiang J., Dong T., Chen Y., Yang X., Liu P. Chelerythrine inhibits the progression of glioblastoma by suppressing the TGFB1-ERK1/2/Smad2/3-Snail/ZEB1 signaling pathway. Life Sci. 2022;293 doi: 10.1016/j.lfs.2022.120358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.