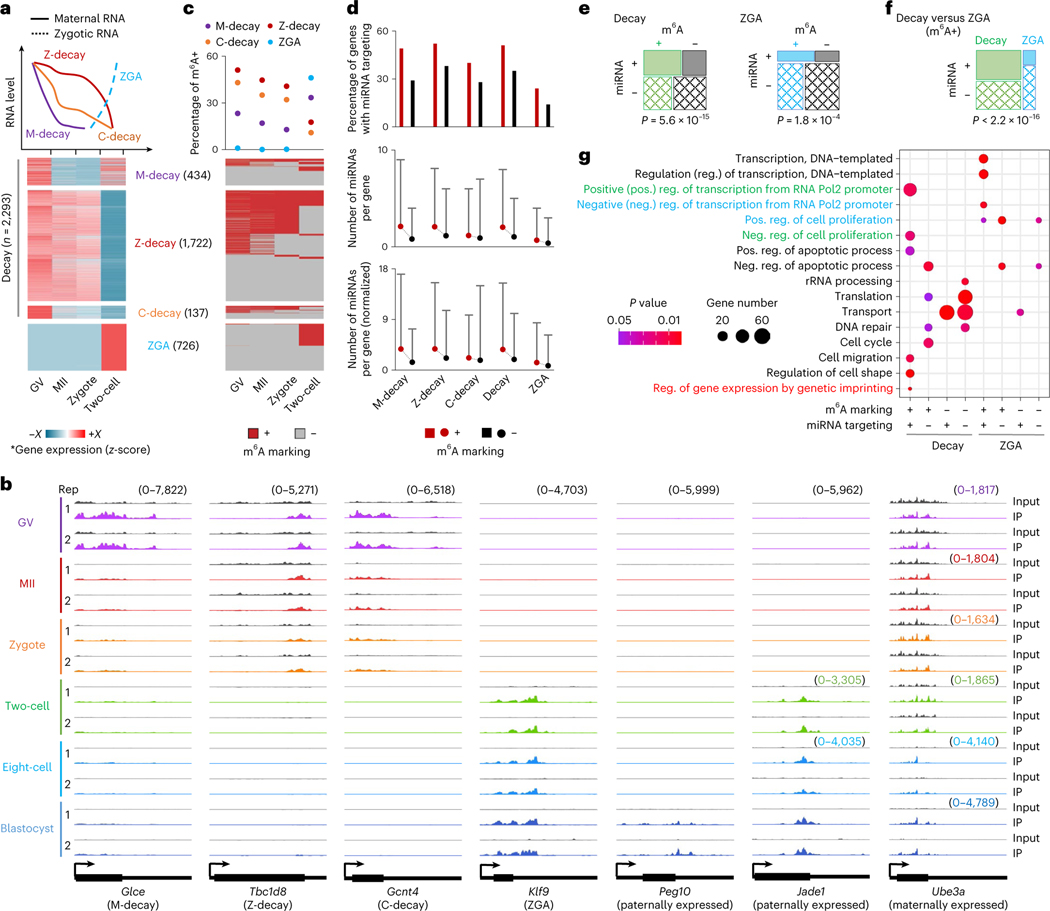
Fig. 3 |. m6A marking and miRNA targeting profiles on Decay and ZGA genes.
a, Definition of Decay and ZGA genes. Top: schematic plot over different expression patterns. Bottom: heat maps of relative gene expression across the four stages. b, Genome browser snapshots of picoMeRIP-seq read density in exonic regions of representative genes. The sample-specific scale ranges are indicated in brackets with the corresponding colors. c, m6A marking status of genes ordered according to a. d, Differential miRNA targeting between m6A+ and m6A− genes. Top: percentage of genes targeted by ≥1 miRNA. Middle: number of miRNAs per gene. Bottom: normalized number of miRNAs per gene, normalized to miRNA expression level. For more details, see Methods. For middle and bottom panels, the dot represents the mean value and the bar is the 95th quantile. e,f, Fisher’s exact test showing the significant miRNA targeting (vertical, ‘+’ for genes that are targeted by miRNA, ‘−’ for genes that are not targeted by miRNA) on m6A+ genes (horizontal, ‘+’ for genes that are marked with m6A, ‘−’ for genes that are not marked with m6A) in Decay and ZGA, respectively (e) and Decay versus ZGA (f). The area of the box represents gene count. For the definition of miRNA targeted genes, see the ‘miRNA analyses’ subsection of Methods. g, GO analyses of genes with different m6A marking and miRNA targeting states in Decay and ZGA genes. Fisher’s exact test was used to calculate the one-sided P values. *The gene expression is log2(TPM + 1), and then normalized by z-score across four stages. The color range (−X to +X): M-decay, −1.72 to 1.73; Z-decay, −1.73 to 1.63; C-decay, −1.44 to 1.73; ZGA, −0.71 to 1.73.