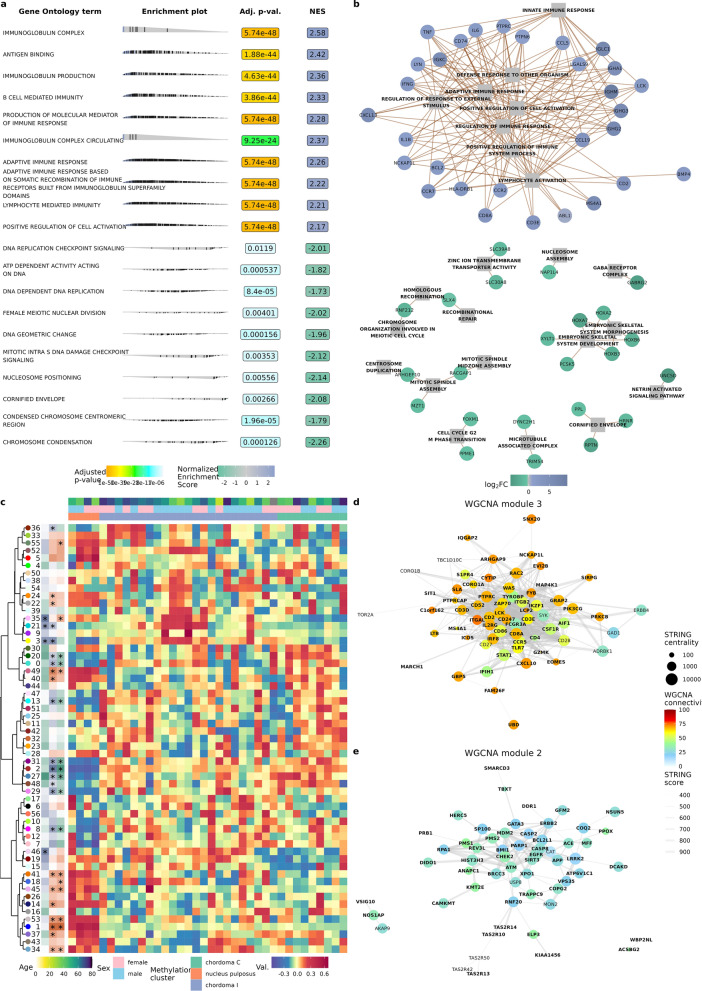
Fig. 3.
Functional analysis of the RNA sequencing results (a) Top 20 terms of gene set enrichment analysis on gene ontology terms (chordoma I–chordoma C comparison) (b) Differentially expressed genes and terms from gene set enrichment analysis. Due to number of terms and genes top hits, basing on p-value, were selected (c) Weighted gene co-expression network analysis, presented on heatmap; samples are shown in columns, WGCNA modules in rows, significance of tests is shown in the panel on the left (d) WGCNA module 3 (with significantly higher score for chordoma I samples) intersected with STRING database; genes to plot were selected based on top connectivity (importance for the module) and centrality (best connected within module); gene names in bold were also differentially expressed in chordoma I, compared with chordoma C (e) Analogous for module 2, higher in chordoma versus nucleus pulposus, DEGs for chordoma—NP comparison are shown in bold